Abstract
Dendritic cells (DC) are the main immune mediators inducing primary immune responses. DC generated from monocytes (MoDC) are a model system to study the biology of DC in vitro, as they represent inflammatory DC in vivo. Previous studies on the generation of MoDC in horses indicated that there was no distinct difference between immature and mature DC and that the expression profile was distinctly different from humans, where CD206 is expressed on immature MoDC whereas CD83 is expressed on mature MoDC. Here we describe the kinetics of equine MoDC differentiation and activation, analysing both phenotypic and functional characteristics. Blood monocytes were first differentiated with equine granulocyte–macrophage colony-stimulating factor and interleukin-4 generating immature DC (iMoDC). These cells were further activated with a cocktail of cytokines including interferon-γ) but not CD40 ligand to obtain mature DC (mMoDC). To determine the expression of a broad range of markers for which no monoclonal antibodies were available to analyse the protein expression, microarray and quantitative PCR analysis were performed to carry out gene expression analysis. This study demonstrates that equine iMoDC and mMoDC can be distinguished both phenotypically and functionally but the expression pattern of some markers including CD206 and CD83 is dissimilar to the human system.
Keywords: antigen presentation, dendritic cells, equine, immunology, microarray
Introduction
Dendritic cells (DC) are the main immune regulators placed at the interface of innate and adaptive immunity. They function as antigen-presenting cells and are the only cells with the ability to induce a primary immune response in naive T lymphocytes.1 In vitro DC systems can be used as models to better understand host–pathogen interactions, for vaccine development and eventually to aid therapeutic protocols. Myeloid DC can be differentiated from peripheral blood monocytes when cultured with granulocyte–macrophage colony-stimulating factor (GM-CSF) and interleukin 4 (IL-4).2,3 The resulting cells are then referred to as monocyte-derived DC (MoDC).3,4 Similar to DC in vivo, monocytes are first differentiated into immature DC (iDC), which are activated to mature DC (mDC) with ‘danger’ signals.2,5 Recent advances in DC biology have highlighted the heterogeneity of DC in vivo and in vitro. In this context it has been suggested that MoDC resemble inflammatory DC in vivo.6 A recent study has demonstrated though that MoDC can substitute for all important functions of DC, including cross-presentation.7 Hence MoDC represent an appropriate model for myeloid DC.
Compared with humans or mice the MoDC system of veterinary animals has not been well characterized but previous studies have shown that MoDC could be generated in various species, including pigs, cattle, sheep, dogs, cats and horses.8–17 However, previous studies failed to demonstrate the clear distinction between iDC and mDC described in humans. In horses, DC were demonstrated in peripheral blood and generated from monocytes.14,17 Equine MoDC were shown to express CD86, MHC II and other markers such as CD11b and CD18. Particularly intriguing was the co-expression of CD206 and CD83 on both equine immature MoDC (iMoDC) and mature MoDC (mMoDC),14 which in humans were used to discriminate immature DC and mature DC. Although lipopolysaccharide (LPS) or poly I : C induced morphological changes in equine MoDC, they did not confer the mDC phenotype previously reported for humans.18–21 This suggested that previous studies had a mixed population or factors in the experimental protocol that influenced differentiation and maturation. We have used a cocktail of pro-inflammatory and anti-inflammatory mediators with the aim of obtaining a more stable mMoDC phenotype.
The key function of DC is the ability to physically interact with and stimulate T lymphocytes. Previous functional studies on equine DC have shown that mMoDC are able to stimulate T cells in in vitro mixed leukocyte reaction assays.14 Here we have taken this a step further by comparing the stimulatory capacity of iMoDC and mMoDC. Other functional attributes of DC, such as endocytosis, phagocytosis and antigen presentation, were also assessed.
In the past, a more comprehensive analysis was hampered by a lack of tools but the sequencing of the equine genome22 has allowed for transcriptomic studies. The changes occurring at the transcriptome in the differentiation and activation states of the equine MoDC system have not been previously investigated. Microarray technology was employed to determine the expression of a broad range of markers for which monoclonal antibodies were not available in the equine system and to analyse the changes in gene expression profiles between monocytes, iMoDC and mMoDC. As a result of the importance of co-stimulatory molecules, such as inducible co-stimulator ligand (ICOS-L), programmed cell death ligand 1 (PD-L1), PD-L2 and B7-H3, in the development of an effective immune response,23 we have assessed the changes in expression of these markers between iMoDC and mMoDC using quantitative real-time PCR. This study demonstrates clear differences in phenotype, function and gene expression between equine MoDC differentiation and activation states.
Materials and methods
In vitro generation of equine monocyte-derived dendritic cells
Peripheral blood mononuclear cells were isolated from healthy horses by Ficoll density centrifugation as previously described.14 Monocytes were further isolated using the monoclonal antibody to human CD14, big 13 clone (Biometec, Greifswald, Germany) also as described elsewhere.24 Monocytes were seeded into 24-well flat-bottom tissue culture plates (Greiner bio-one, Stonehouse, UK) at a concentration of 2 × 106 cells per well in 1 ml RPMI-1640 medium (Gibco-Invitrogen, Paisley, UK) supplemented with 10% fetal calf serum (Autogen Bioclear, Calne, Wiltshire, UK), 0·1 mg/ml of penicillin and streptamycin (Gibco-Invitrogen) and 2% HEPES (Gibco-Invitrogen). Cells were differentiated with the addition of 1000 and 500 U/ml of purified recombinant equine GM-CSF and IL-4, respectively and cultured for 5 days. Before stimulation, the bioactivity of GM-CSF and IL-4 was quantified as 8 × 107 and 1 × 105 U/ml, respectively using the human TF-1 cell (ECACC, Salisbury, UK) proliferation assay as previously described14 and subsequently titrated on equine monocytes to adjust for species differences. For maturation, dendritic cells were exposed to 1 μg/ml LPS and 20 μg/ml poly I : C or a DC maturation cocktail comprising 20 ng/ml equine tumour necrosis factor -α (R&D Systems, Abingdon, UK) 10 ng/ml equine IL-1β (R&D Systems), 20 μg/ml equine IL-6 (R&D Systems), 1 μg/ml prostaglandin E2 (Enzo Life Sciences, Exeter, UK) and 100 ng/ml equine IFN-γ (R&D Systems). All reagents, such as media, FCS, Ficoll, recombinant cytokines and maturation stimuli were tested to exclude LPS contamination.
Analysis of cell surface marker expression
To analyse the expression of surface markers, cells were stained with the live/dead fixable violet dead cell kit (Invitrogen, Paisley, UK) and analysed using anti-human CD14 monoclonal antibody big 13 (Biometec), anti-human CD206 clone 3.29B1.10 (Beckman Coulter, High Wycombe, UK), anti-human CD83 clone HB15a (Beckman Coulter), anti-human CD86 clone IT2.2 (Becton Dickinson, Oxford, UK) and an anti-horse MHC II clone EqT2 (VMRD, Pullman, WA). Some antibodies were not directly labelled and were either labelled via the zenon kit (Invitrogen) or indirectly labelled. Analysis was performed according to previously described protocols.25 Stained cells were analysed using a MACSQuant Analyzer and MACSQuant software (Miltenyi Biotec, Bergisch Gladbach, Germany). Statistical analysis here and for other assays was performed using graphpad prism 5 software.
Functional assays
Endocytic and phagocytic assays
The ability of MoDC to endocytose allophycocyanin-labelled ovalbumin (OVA-APC; Fisher Scientific, Leicestershire, UK) or phagocytose FITC-labelled FluoSphere carboxylate-conjugated microsphere particles (1·0-μm diameter; Invitrogen) was assayed by flow cytometry following previously published protocols.17,26,27 Briefly, freshly isolated monocytes, iMoDC or mMoDC were washed once and resuspended in RPMI-1640 medium at a density of 1 × 105 cells per well of a flat-bottomed 96-well plate (Invitrogen). All plates were incubated on ice for 30 min before adding OVA-APC to a final concentration of 20 μg/ml and FITC-conjugated carboxylate-modified microspheres at a ratio of 5 : 1 (beads/cell). Cells were incubated at 4° (control) and 37° for 1 hr or 4 hr for the endocytic and phagocytic assays respectively, subsequently washed three times with cold PBS solution (Invitrogen) and re-suspended in PBS for flow cytometric analysis.
Mixed leucocyte reactions
Equine T lymphocytes were enriched using anti-horse CD5, clone CVS5 (Serotec, Kidlington, UK) indirectly labelled to anti-mouse IgG microbeads (Miltenyi Biotec) and magnetically sorted. The MoDC from one horse were added in graded doses to 5 × 105 CFSE-labelled T lymphocytes from another horse. The protocol for labelling of cells with CFSE was carried out as previously described.28 Subsequently, cells were co-cultured at 37° for 3 days, before proliferation of T cells was measured by flow cytometry as previously described.7 Cells were harvested, washed twice using PBS with 10% fetal calf serum and stained with the live/dead fixable violet dead cell kit to exclude all dead cells from analysis.
Antigen presentation
Graded numbers of iMoDC were incubated at 37° for 2 hr with 1 mg/ml LPS-free OVA, which can be considered an antigen that horses do not encounter. After incubation, iMoDC were matured overnight with the cocktail as described before. CFSE-labelled T lymphocytes from the same horse were added to the mMoDC at a density of 5 × 105 cells and co-cultured at 37°. After 4 days, proliferation of live T cells was evaluated by flow cytometry as in the mixed leucocyte reaction assays.
To determine the ability of MoDC to cross-present antigen, the protocol was similar as described above. However, autologous CFSE-labelled CD8+ T cells were magnetically sorted and added to mMoDC in a DC : T-cell ratio of 1 : 10 and co-cultured at 37° for 5 days. Controls included mMoDC only, mMoDC in the presence of either OVA or T cells and concanavalin A-stimulated T cells in the presence of OVA. Cells were stained with anti-horse CD8 conjugated to Alexa Fluor 700 APC via the zenon labelling kit to define T cells in the analysis.
RNA extraction and microarray
Total RNA extraction was performed using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany), following the manufacturer's instructions. RNA was quantified using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA quality was assessed with the RNA Nano or Pico 6000 Labchip kit on the Agilent 2100 Bioanalyzer (Agilent Technologies, Berkshire, UK). Microarray experiments were performed following the Agilent one-colour gene expression system and the horse catalogue array (Agilent Design ID 021322). Briefly, target RNA was amplified and labelled for the generation of complementary RNA using the Low Input Quick Amp Labelling Kit (Agilent Technologies). Samples were hybridized to the Gene Expression Microarray (Agilent Technologies) and washed following the protocol of the Gene expression hybridization kit (Agilent Technologies). Three biological repeats were analysed for each data set. The arrays were scanned with the Agilent High-resolution C Microarray Scanner and the raw data were extracted with Agilent's Feature extraction software. Data quality was assessed by the specific quality control reports of metrics targeted to the experiment. All data analysis was performed in GeneSpring gx software version 11.5.1 (Agilent Technologies).
The raw data were pre-processed by log2 transformation followed by scale normalization. The parametric statistical test of analysis of variance (anova) unequal variance (Welch anova) was used to test differential expression between monocytes and iMoDC, where monocytes were used as the reference, and differential expression between iMoDC and mMoDC, where iMoDC were used as the reference. Benjamini Hochberg test was used to correct for multiple testing (false discovery rate of 0·05). The threshold of significance was set to a minimum fold change of 2. Unsupervised hierarchial clustering on both probes and cell types was performed to identify patterns within the data sets using the Euclidean similarity metric and Hierarchial clustering algorithm method with the Centroid linkage rule. The output differential gene expression lists were curated by eliminating all un-annotated EST sequences. The gene symbols were assigned to each probe based on the Agilent probe descriptions. Principal component analysis was performed on the curated differential expression gene lists to assess differences in expression profiles between cell types.
Quantitative real-time PCR
A selected set of co-stimulatory genes was used to validate the results of microarray (PDL1/CD274, PDL2/CD273 and B7-H3/CD276) and to expand the results for genes not on the array (ICOSL/CD275). Equine-specific primers were designed with Primer329 and primer sequences are shown in Table 1. Synthesis of cDNA was performed with the SuperScript II First-Strand Synthesis System using random hexamer primers (Invitrogen). Real-time quantitative PCR was performed in triplicate each with a 25-μl final reaction volume containing 400 nm of each primer, 1 μm of probe and 5× Quantitect PCR Master Mix with ROX reference dye (Qiagen). An 18S gene quantitative PCR was used as the endogenous control in all samples30 (Applied Biosystems, Foster City, CA). The thermal profile consisted of a denaturation step at 95° for 10 min followed by 40 cycles at 95° for 15 s and 60° for 1 min. The PCR was analysed by relative quantification using the △△Ct method.31 Statistical analysis was performed using graphpad prism 5 software.
Table 1.
Primer and probe sequences used to measure surface marker expression of the co-stimulatory molecules
| Gene | Primer and probe sequence 5′→3′ | Accession no. reference sequence |
|---|---|---|
| PD-L1/ | TGGTGGTGCTGACTACAAGC1 | Genbank: XM_001492842 |
| CD274 | GTGGTCACTGCTTGTCCAGA2 | |
| 6FAM-ATTTCTGTGGATCCGGTCAC-BHQ-13 | ||
| PD-L2/ | CTTTGGATGACCCAGCACTT1 | RefSeq: XM_001492097 |
| CD273 | AAGGAGCCTCAGGACACTCA2 | |
| 6FAM-TGTGCTCAAAGGAAGTCAGGC-BHQ-13 | ||
| ICOSL/ | TCCAAGGCCGAATGTCTACT1 | RefSeq: C_009169 |
| CD275 | GCACGTTCTCTATGCAGCAG2 | |
| 6FAM-TCAACAAGACGGACAACAGC -BHQ-13 | ||
| B7-H3/ | AATCAGACCATCCAGCGTGT1 | RefSeq: XM_001493661 |
| CD276 | GAGGCAGAACCACAGCACTC2 | |
| FAM-GAGAGCCAGCTGTCAGCTG-BHQ-13 |
Forward primer sequence.
Reverse primer sequence.
Probe sequence.
Results
Equine MoDC co-express CD83 and CD206
In contrast to studies with human cells, a previous study with equine MoDC had demonstrated that CD206 was not necessarily expressed on all equine MoDC. As the induction of CD206 has been shown to be dose-dependent on IL-4, we first determined the optimal dose of IL-4 to induce CD206 on equine monocytes. We could indeed confirm a dose-dependent expression of CD206. However, at best only around half of all the monocytes responded to equine IL-4 with an induction of CD206, which was stably induced above 200 U/ml (Fig. 1). This was in contrast to the human system where a lower concentration of IL-4 maximally induced the expression of CD206 on around three-quarters of monocytes.32
Figure 1.
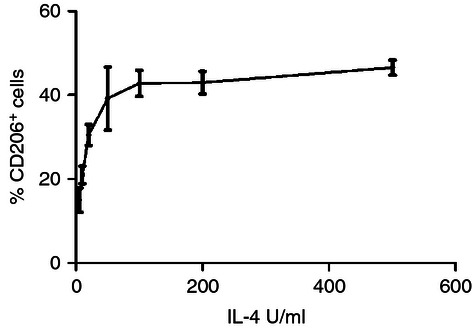
Effect of interleukin-4 (IL-4) on the expression of CD206. Monocytes were cultured in the presence of increasing concentrations of IL-4. After 2 days, the cells were stained with anti-CD206 and assessed by flow cytometry. Data are represented as the mean percentage positive cells ± SEM (n = 3).
Another intriguing finding of a previous study on equine MoDC was the early co-expression of CD206 and CD83,14 considered markers of human iMoDC and DC maturation in humans, respectively.33–35 Having determined 500 U/ml as an ample concentration for equine monocytic cells to express CD206, we studied the kinetics of CD206 and CD83 during MoDC differentiation. Early-stage iMoDC quickly expressed mainly CD206 but gradually became double positive for CD206 and CD83. Late-stage iMoDC showed CD83+ only cells before any activation signal (Fig. 2a). Subsequently, we compared several activation stimuli such as LPS, poly I:C and a cocktail of factors as described previously36,37 including soluble CD40 ligand and equine interferon-γ. The maturation cocktail clearly induced the best activation of mMoDC with a significant up-regulation of MHC II, CD86 and CD83 while down-regulating CD206 (Fig. 3). However, next to the majority of CD83+/CD206− cells, mMoDC still possessed a minor population of cells co-expressing both markers along with a very small percentage of cells expressing only CD206 (Fig. 2b).
Figure 2.
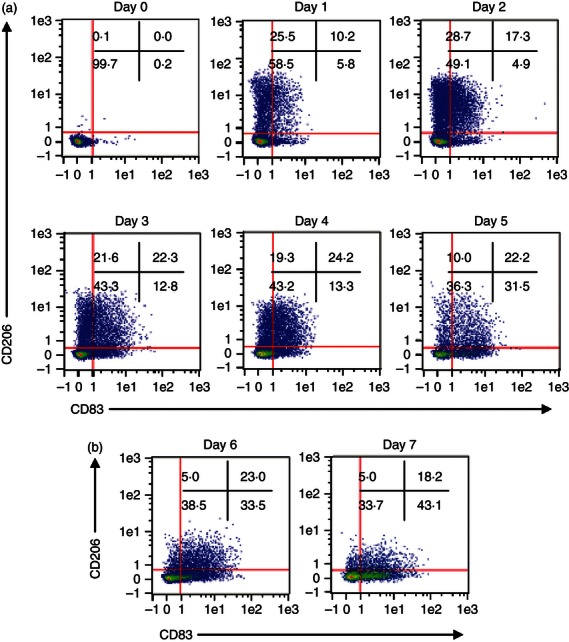
Co-expression of CD206 and CD83 on equine monocyte-derived dendritic cells (MoDC). Monocytes were cultured in the presence of 1000 U/ml granulocyte–macrophage colony-stimulating factor (GM-CSF) and 500 U/ml interleukin-4 (IL-4) for 5 days then activated with a cocktail of inflammatory mediators. The cells were harvested daily and stained with phycoerythrin (PE) -conjugated anti-human CD206 clone 3.29B1.10 and PE-Cy5 conjugated anti-human CD83 clone HB15a for 30 min at 4° and analysed by flow cytometry. Live-dead staining was performed on all cultures, using a fixable violet dead cell stain kit, to exclude dead cells from the analysis. The flow cytometry dot plots show the relationship between CD206 and CD83. The expression of CD206 was high on early-stage immature MoDC with cells gradually becoming double positive for CD206 and CD83. The late stage immature MoDC and mature MoDC showed an increase in CD83 expression but still maintaining the co-expression of CD206 and CD83. Data are representative of three independent repeats.
Figure 3.
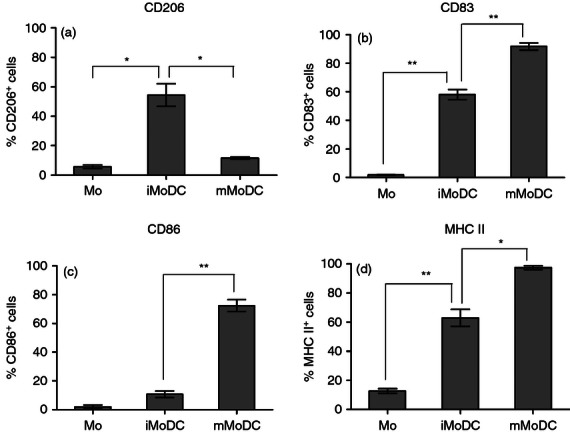
Phenotypic analysis of equine monocyte-derived dendritic cell (MoDC). Monocytes were stimulated with equine cytokines for 5 days then matured with a cocktail comprising equine tumour necrosis factor-α (TNF-α), equine interleukin-1b (IL-1β), eqIL-6, prostaglandin E2 (PGE2) and equine interferon-γ (IFN-γ) for 48 hr. The bar graphs compare the expression of the main DC surface markers on monocytes, immature MoDC (iMoDC) and mature MoDC (mMoDC). (a) The percentage cells expressing mannose receptor (CD206) increased during differentiation but reduced upon maturation. (b) CD83 showed minimal expression on monocytes but was highly expressed on iMoDC and increased further on mMoDC. (c) The expression of CD86 was low on both monocytes and iMoDC but increased on mMoDC. (d) MHC II was highly expressed on iMoDC and increased on mMoDC. The number of positive cells corrected for isotype control was obtained from the main fluorescence channel using MACSQuant software and represents the average mean ± SEM (n = 3). For single comparisons between Mo→iMoDC and iMoDC→mMoDC, a two-tailed paired Student t-test was used. * and ** indicate significant differences between sample means P < 0·05 and P < 0·01, respectively.
Equine iMoDC and mMoDC have distinctly different functional attributes
More important than phenotypical profiles, which may vary largely between DC, are functional parameters that define DC. Whereas monocytes possessed the highest ability to endocytose OVA-APC (Fig. 4a), iMoDC exhibited a more potent endocytic and phagocytic capacity than mMoDC (Fig. 4a,b), hence matching the expectation for DC.
Figure 4.
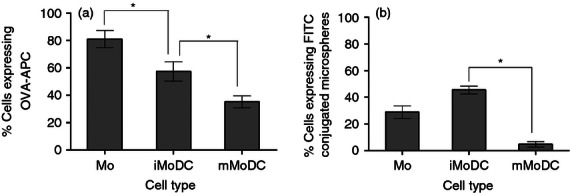
The capacity of equine monocyte-derived dendritic cells (MoDC) to take up antigen. (a) Effect of stimulation and activation on equine dendritic cells endocytic capacity. Mo, immature MoDC (iMoDC) and mature MoDC (mMoDC) were incubated with 20 μg/ml of ovalbumin (OVA) at 4° and 37° for 1 hr. The percentage of cells expressing allophycocyanin-conjugated OVA (OVA-APC) represent the difference in values obtained between the 37° and 4° (control). Monocytes have the greatest ability to take up OVA compared with DC while iMoDC are better at uptake than mMoDC. (b) Effect of stimulation and activation on equine DC phagocytic capacity. Immature MoDC and mMoDC wrere incubated at 4° and 37° for 1 hr with an FITC-conjugated microsphere,1·0 μm in size, at a 5 : 1 (beads/cell) ratio. Immature MoDC compared with mMoDC have a higher phagocytic activity. There was no significant difference between Mo and iMoDC. The percentage of positive cells represents the average mean ± SEM (n = 3). For single comparisons between Mo→iMoDC and iDC→mMoDC, a two-tailed paired Student's t-test was used. * indicate significant differences between sample means P < 0·05.
Cultured iMoDC and mMoDC were further examined for their ability to stimulate allogeneic T cells in a primary mixed leucocyte response assay. As shown in Fig. 5(a), mMoDC showed a high allostimulatory potential whereas equine iMoDC are relatively modest stimulators compared with mMoDC.
Figure 5.
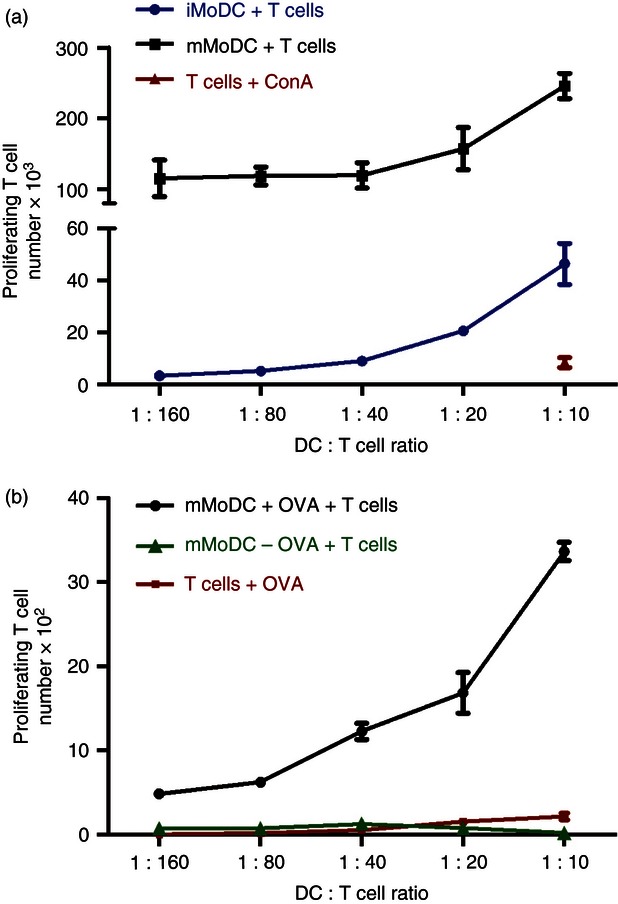
The ability of equine monocyte-derived dendritic cells (MoDC) to stimulate T cells and present antigen. (a) Comparative ability of immature MoDC (iMoDC) and mature MoDC (mMoDC) stimulation of allogeneic T cells. MoDC were incubated with CFSE-labelled allogeneic T cells at 37° for 3 days. Cells were then harvested and stained with an anti-equine CD5 antibody to gate T cells and a fixable violet dead cell stain to exclude dead cells from the analysis. Samples were run on the MACSQuant flow cytometer. Mature MoDC have a higher T-cell activation potential than iMoDC. (b) Presentation of ovalbumin (OVA) to autologous T cells by graded doses of MoDC. The iMoDC were incubated for 2 hr with 0·02 mg/ml of OVA, then matured with the cocktail of inflammatory mediators and left overnight. After which CFSE-labelled autologous T cells were added to cultures and incubated at 37° for 5 days. MoDC have the ability to present antigen to autologous T cells. Proliferating T-cell numbers represent the average mean ± SEM (n = 3).
To test the capacity of equine DC to present exogenous proteins, iMoDC were incubated with soluble ovalbumin before maturation and the addition of autologous T cells. Figure 5(b) shows the ability of equine MoDC to induce T-cell proliferation in the presence of exogenous protein OVA in a primary stimulation.
Equine MoDC were also assessed for their cross-presentation ability. The MoDC were able to cross-present OVA to autologous CD8+ T cells, thereby inducing proliferation (Fig. 6). As the purity of CD8+ T cells after magnetic isolation was over 95% and controls failed to elicit a response, this confirms that equine MoDC fully resemble functional DC.
Figure 6.
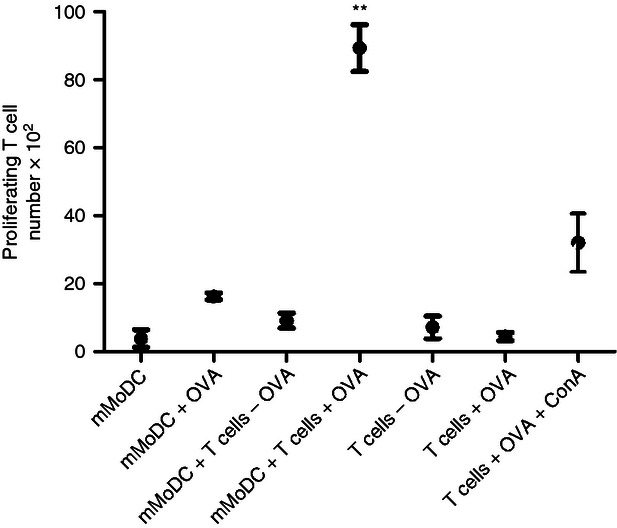
Presentation of soluble antigen to CD8+ T cells by mature monocyte-derived dendritic cells (mMoDC). Immature MoDC were incubated for 2 hr with 0·02 mg/ml ovalbumin (OVA), and then matured. Autologous CD8+ T cells were magnetically sorted with anti-horse CD8 antibody (clone CVS8) conjugated to anti-mouse IgG microbeads, CFSE-labelled, added to cultures in a DC : T-cell ratio of 1 : 10 and incubated at 37° for 5 days. MoDC have the ability to cross-present antigen to CD8+ T cells. Proliferating T-cell numbers represent the average mean ± SEM (n = 3). For single comparisons between mMoDC + T cells + OVA and mMoDC + T cells – OVA, a two-tailed paired Student's t-test was used. ** indicates a significant difference between sample medians where P is < 0·005.
Gene expression analysis
Although the above results demonstrate the differentiation and activation of equine MoDC, the lack of antibodies in the horse system prevents the performance of a more comprehensive analysis. We therefore resorted to gene expression profiling using a commercially available equine-specific microarray. Expression profiles of all three cell types displayed by 3D Principal Component Analysis (Fig. 7a) showed that the three cell types are indeed distinct populations. The relationship between monocytes, iMoDC and mMoDC was also examined by unsupervised hierarchial clustering. Here, the distinction of the three cell types was further confirmed with the heat map and revealed that iMoDC and mMoDC are closer to each other than to monocytes, but differences in their gene expression profiles make them clearly distinct populations (Fig. 7b).
Figure 7.
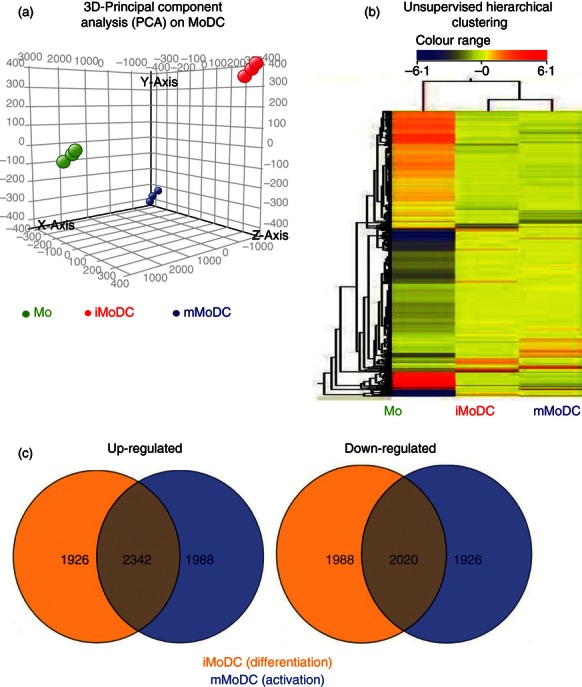
Gene expression analysis of equine monocyte-derived dendritic cells (MoDC). Microarray experiments were performed using the Agilent Horse Gene Expression Array system (4 × 44 K). This array design is based on the first draft sequence of the horse genome (Agilent design ID 021322 released 2009). All data sets represent three biological repeats and were analysed using GeneSpring. (a) 3D-Principal component analysis (PCA) performed on the differentially expressed genes in monocytes, immature MoDC (iMoDC) and mature MoDC (mMoDC) shows that their gene expression profiles are different and therefore are three distinct cell types. (b) Heat map by unsupervised hierarchical clustering of the differentially expressed probes between monocytes, iMoDC and mMoDC. The iMoDC and mMoDC are closer to each other than to monocytes, but the expression profiles of the three cell types are clearly segregated indicating differences in the total RNA expression pattern in Mo and DC. Red represents up-regulation, blue down-regulation and yellow represents no change for differentially expressed genes. (FDR < 0·05, fold change ≥ 2·0). (c) Venn diagram representing the number of probes up-regulated and down-regulated in the differentiation and activation states. There were over 4000 probes differentially expressed in iMoDC and mMoDC. A total of 1926 probes were up-regulated in iMoDC only and 1988 in mMoDC whereas 1988 probes were down-regulated only in iMoDC and 1926 only in mMoDC.
An unpaired standard t-test (P ≤ 0·01) provided a list of 8268 and 8276 differentially expressed probes for the differentiation and activation states, respectively. The numbers of probes significantly up-regulated and down-regulated in iMoDC only were 1926 and 1988, respectively. Whereas 1988 and 1926 probes were up-regulated and down-regulated, respectively in mMoDC only. However, 2342 and 2020 probes up-regulated and down-regulated, respectively were common to both iMoDC and mMoDC (Fig. 7c). However, as the gene lists were curated by eliminating all un-annotated EST and cDNA library sequences and averaging the mean fold change of repeat probes, a final list of 526 genes, differentially expressed between Mo and iMoDC, and 535 genes, differentially expressed between iMoDC and mMoDC, was obtained.
The array confirmed the already high expression of CD83 on iMoDC, which suggests that maturation in the equine system is not linked to CD83 expression as in the human system. Within the differentially expressed genes, we then assessed the expression of the co-stimulatory B7 family ligands. Similar to CD86 (Fig. 3), CD80 was expressed on iMoDC but further up-regulated on mMoDC. The co-stimulatory molecules CD273/PD-L2, CD274/PD-L1, CD275/ICOS-L and CD276/B7-H3 were also analysed using TaqMan-based real-time PCR assays (Fig. 8). All co-stimulatory molecules were up-regulated on iMoDC during differentiation. Upon activation, the expression of ICOS-L and B7-H3 remained stable on mMoDC, while PD-L1 and PD-L2, which are negative regulators of immunity, were down-regulated. These data confirm the microarray analysis for PD-L1, PD-L2 and B7-H3 and add valuable information for ICOS-L, as this gene was not represented on the array.
Figure 8.
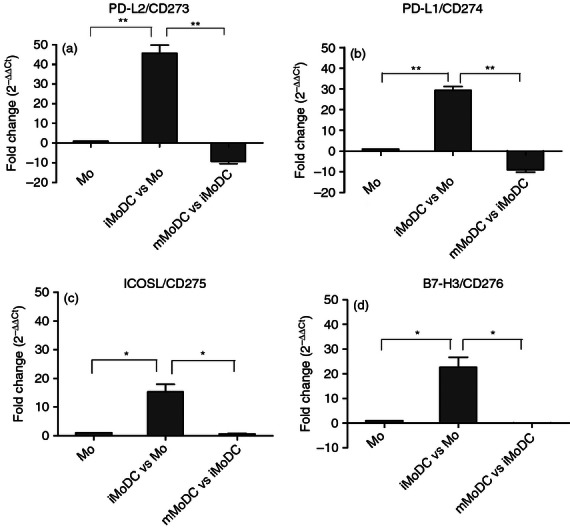
Validation of differentially expressed co-stimulatory molecules by TaqMan real-time quantitative PCR. The B7 co-stimulatory molecules were analysed using TaqMan based real-time PCR. The expression of these co-stimulatory molecules was up-regulated on immature monocyte-derived dendritic cells (iMoDC) and down-regulated (PD-L1/CD274 and PD-L2/CD273) or unchanged (ICOS-L/CD275 and B7-H3/CD276) on mature MoDC (mMoDC). Results represent the average fold change ± SEM (n = 3). For single comparisons between Mo→iMoDC and iMoDC→mMoDC, a two-tailed paired Student's t-test was used. * and ** indicate significant differences between sample means P < 0·05 and P < 0·01, respectively.
Chemokine receptors and their ligands are involved in the migration of DC and reflect their differentiation and activation states.38,39 CCR7 was induced on iMoDC and further up-regulated on mMoDC (Table 2). Molecules downstream in the signalling cascade of CCR740 were also differentially expressed in equine MoDC (Table 2). CCR5 was up-regulated on iMoDC and further up-regulated during maturation. The expression of other chemokines was similar or varied between the differentiation and activation states. Chemokines CCL17/TARC, CXCL13/BCA-1 and CCL2/MCP-1 were constitutively expressed by iMoDC and mMoDC, whereas chemokines CXCL9/Mig, CXCL11/I-TAC and CXCL10/IP-10 were negatively modulated or low on iMoDC but positively modulated on equine mMoDC (Table 2).
Table 2.
A selection of genes differentially expressed during differentiation and upon activation of equine monocyte-derived dendritic cells (MoDC). Total RNA analysis of immature and mature MoDC was performed by microarray. To determine the differentially expressed genes during differentiation and upon activation, Mo and immature MoDC were used as the reference samples, respectively. The threshold of significance was set to a minimum fold change of 2
| Fold change | |||
|---|---|---|---|
| Gene symbol | Differentiation Mo→iMoDC | Activation iMoDC→mMoDC | |
| CD83 | 12·7 | 8·0 | |
| Co-stimulatory molecules | PD-L1/CD274 | 56·0 | −15·3 |
| PD-L2/CD273 | 42·0 | −12·9 | |
| B7-H3/CD276 | 21·5 | −2·3 | |
| B7-1/CD80 | 2·1 | 3·5 | |
| Dendritic cell migration | CCR7 | 1137·5 | 2·3 |
| Rho | 2·29 | 19·1 | |
| CCR5 | 7·7 | 2·1 | |
| Chemokines | CCL17/TARC | 13579·9 | −2·7 |
| CXCL13/BCA-1 | 479·36 | −18·9 | |
| CCL2/MCP-1 | 116·0 | −2·0 | |
| CXCL9/Mig | −3·0 | 726·8 | |
| CXCL11/I-TAC | 3·6 | 120·8 | |
| CXCL10/IP-10 | −6·4 | 32·5 | |
Discussion
Purified recombinant cytokines were used to drive the controlled differentiation of equine MoDC. Under such stringent conditions a highly reproducible system to obtain iMoDC, which could be matured into mMoDC through a cytokine cocktail, could be established.
While phenotypic data demonstrated the modulation of key markers, the expression patterns of CD83, CD206 and to some extent MHC II are clearly not in agreement with what has been described in humans or mice. CD83 is considered to be a marker of maturation on human and murine DC19,41 but it is already expressed on equine iMoDC. Previous studies in humans and mice have shown that CD83 is correlated with the density of MHC II on antigen-presenting cells and conversely a lack of CD83 on mature DC is associated with their inability to stimulate T cells during mixed leucocyte reaction.42–44 It is therefore not surprising that MHC II was also co-expressed on equine iMoDC and the ability of iMoDC to already stimulate T cells is correlated to the MHC II and CD83 expression on these cells.
Murine MoDC were shown in vitro to be as good as, or better than, classical DC at cross-presentation.7 Here, it is demonstrated that equine MoDC also possess the ability to cross-present antigen, which in myeloid DC is otherwise a particular attribute of DNGR1/CLEC9A (C-type lectin-like domain family 9) -positive DC45,46 the expression of which could not be determined here because of a lack of antibodies cross-reacting. Overall, this emphasizes the capacity of MoDC as suitable tools for ex vivo immunotherapy approaches.47–50
Similarly, the expression of CD206 has been the hallmark of immature DC differentiation in humans, not expressed on monocytes or mature DC and its link to receptor-mediated endocytosis by DC has been well described.18,20,21 Mature MoDC still expressing CD206 in the equine system were probably the cells conveying the remaining capacity to endocytose antigen. The spontaneous transition of iMoDC to mMoDC could be excluded in our system because both phenotypical and functional studies clearly demonstrated differences between equine iMoDC and equine mMoDC whereas sufficient similarities to human and mouse immature and mature DC exist to classify them as such.
To obtain fully activated MoDC in vitro, we assessed different maturation stimuli, including LPS, poly I:C, a cocktail of inflammatory cytokines and the cocktail in combination with interferon-γ or CD40 ligand. Our data support the previously published notion14 that LPS or poly I:C were inefficient at driving maturation (not shown) and are in line with a recent comparison on human mature DC.37 Our data contrast with those of a previous study stimulating equine DC with inactivated Escherichia coli where no difference in the CD206-mediated endocytic capacity of unstimulated and activated equine DC could be detected.17 This highlights the importance of using an appropriate activation stimulus to obtain a robust maturation.
To obtain further insights into the differentiation and activation status of equine MoDC, gene expression studies were applied where antibodies were not available for horses. Microarray data such as principal components analysis and hierarchical clustering underpinned the fact that equine iMoDC and mMoDC are two distinct stages.
Co-stimulatory molecules were generally regulated along the function of equine MoDC. The expression of B7-1/CD80 was low on equine iMoDC but up-regulated on equine mMoDC, which is comparable to the expression on human MoDC.14 PD-L1/CD274 and PD-L2/CD273 were up-regulated on equine iMoDC but upon activation were down-regulated, but still expressed (Fig. 7). On human DC PD-L1 and PD-L2 are significantly up-regulated on mMoDC.51 Both can suppress the immune system by transmitting an inhibitory signal, which negatively regulates T-cell activation,52–55 but have also been reported to stimulate T-cell proliferation.56,57 The down-modulation of these markers on equine mMoDC implies an inhibitory role in the equine immune responses, which upon activation would not be intended at first. While ICOS-L/CD275 is expressed at low levels on human monocytes and remains unaltered during differentiation and maturation of MoDC,58 differentiation of equine MoDC strongly induced ICOS-L expression, which was sustained during maturation. ICOS-L (CD275) is a positive co-stimulatory signal for T cells, which drives the production of IL-10 in T cells and seems to be particularly relevant to the induction of T helper type 2 (Th2) cells.59,60 The expression pattern of B7-H3/CD276 is similar to human MoDC, up-regulated on equine iMoDC and stable on equine mMoDC.61 B7-H3 was reported to be involved in T-cell activation62,63 but further studies suggested that it may negatively regulate T cells.64–66 The activity of equine MoDC argues against an inhibitory role of B7-H3 on equine MoDC.
To gain some insight into the migratory ability of equine MoDC, we used the results from the microarray to analyse the expression of chemokines and their receptors in more detail. These molecules were some of the most highly regulated genes and indicate the ability of equine MoDC to interact with other cells. CCR7 is key for migration of DC toward T-cell areas but is already strongly expressed on equine iMoDC, whereas in the human system its expression is mostly up-regulated during maturation.38,67–69 The chemokine receptor CCR5, which binds to ligands such as regulated on activation, normal T-cell expressed and secreted (RANTES)/CCL5, macrophage inflammatory protein-1α (MIP-1α)/CCL3 and MIP-1β/CCL4, was up-regulated during differentiation and activation of equine MoDC. In contrast, human MoDC38,68,70 have been reported to down-regulate CCR5 and to lose their responsiveness to its ligands upon maturation.38,68 RANTES and MIP-1β are secreted by T lymphocytes,71 so the expression of CCR5 may support the interaction of equine mature DC and T cells.
Chemokine production by DC enhances their capacity to attract other cells. CCL17/TARC, CXCL13/BCA-1 and CCL2/MCP-1 were all highly regulated during differentiation and remained expressed. CCL17, one of the ligands for CCR4, was the most highly regulated chemokine detected and has a selective activity towards Th2 cells.38,72,73 Further studies will be required to establish if this negatively impacts the ability of equine MoDC to initiate a Th1 response. CXCL13, a ligand for CXCR5, has been implicated in establishing the interaction of DC with T and B cells, which specifically suits the function of mature DC.74 CCL2 has been shown to inhibit IL-12 production and promote Th2 polarization, also indicating a balance of equine MoDC towards Th2.75,76
The expression of chemokines CXCL9/Mig, CXCL11/I-TAC and CXCL10/IP-10, all ligands for receptor CXCR3, was specifically up-regulated upon activation. Human DC expressing high levels of these chemokines have been shown to attract CD8+ T cells expressing the CXCR3 receptor.77 Their high expression on equine mMoDC may suggest that these cells are also efficient at attracting CD8+ T cells, and the cross-presenting ability of these cells supports an interaction of DC and CD8+ cells.
As indicated here, transcriptome analysis may contribute substantially to our understanding of the differentiation and maturation of equine DC. It needs to be mentioned, however, that this first-generation equine array resembled only part of the equine RefSeq database and was poorly annotated. Further work is necessary to exploit the advancements in equine genomics.
In summary, it has been demonstrated here that equine iMoDC and equine mMoDC are distinct cell populations, where neither CD83 nor CD206 are correlated with differentiation or maturation and cannot be used to distinguish stages of eqMoDC. Although this opposes studies on human MoDC, it complements previous studies in the equine system14,16,17 but is similar to the situation described in other veterinary species, where markers such as CD83 or CD206 were not best placed to discriminate immature from mature DC either and danger signals like LPS or tumour necrosis factor-α alone did not convey full maturation.78,79
The equine MoDC system presented is robust and can be used to further investigate Mo and DC subpopulations and their function, for example when encountering pathogens. Further work will be necessary to investigate if equine DC are in line with the proposed pan-species classification of DC.80
Acknowledgments
We thank Dr Ute Weyer of the Animal Service Unit (ASU), Animal Health and Veterinary Laboratories Agency (AHVLA), for assistance with the horses. This work was supported by the AHVLA SC0191 project. We also thank Dr Ernesto Oviedo-Orta (University of Surrey) who supervised the PhD project part associated to this manuscript.
Glossary
- MoDC
monocyte-derived dendritic cells
- Eq
equine
- iMoDC
immature MoDC
- mMoDC
mature MoDC
Disclosure
The authors have no financial conflict of interest.
References
- 1.Steinman RM. The dendritic cell system and its role role in immunogenicity. Annu Rev Immunol. 1991;9:271–96. doi: 10.1146/annurev.iy.09.040191.001415. [DOI] [PubMed] [Google Scholar]
- 2.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocytes/macrophage colony stimulating factor plus interleukin 4 and downregulated bt tumor necrosis factor α. J Exp Med. 1994;194:1109–18. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peters JH, Gieseler R, Thiele B, Steinbach F. Dendritic cells: from ontogenic orphans to myelomonocytic descendants. Immunol Today. 1996;17:273–8. doi: 10.1016/0167-5699(96)80544-5. [DOI] [PubMed] [Google Scholar]
- 4.Steinbach F, Kruse B, Thiele B. Monocyte derived dendritic cells (MODC) present phenotype and functional activities of Langerhans cells/Dendritic cells. Adv Exp Med Biol. 1995;2:151–3. doi: 10.1007/978-1-4615-1971-3_33. [DOI] [PubMed] [Google Scholar]
- 5.Thery C, Amigorena S. The cell biology of antigen presentation in dendritic cells. Curr Opin Immunol. 2001;13:45–51. doi: 10.1016/s0952-7915(00)00180-1. [DOI] [PubMed] [Google Scholar]
- 6.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–61. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheong C, Matos I, Choi JH, et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209+ dendritic cells for immune T cell areas. Cell. 2010;143:416–29. doi: 10.1016/j.cell.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howard CJ, Brooke GP, Werling D, Sopp P, Hope J, Parsons KR, Collins RA. Dendritic cells in cattle: phenotype and function. Vet Immunl Immunopathol. 1999;72:119–24. doi: 10.1016/s0165-2427(99)00124-5. [DOI] [PubMed] [Google Scholar]
- 9.Werling D, Hope JC, Chaplin P, Collins RA, Taylor G, Howard CJ. Involvement of caveolae in the uptake of respiratory syncytial virus antigen by dendritic cells. J Leukoc Biol. 1999;66:50–8. doi: 10.1002/jlb.66.1.50. [DOI] [PubMed] [Google Scholar]
- 10.Carrasco C, Rigden R, Schaffner R, et al. Porcine dendritic cells generated in vitro: morphological, phenotypic and functional properties. Immunology. 2001;104:175–84. doi: 10.1046/j.0019-2805.2001.01299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paillot R, Laval F, Audonnet J, Andreoni C, Juillard V. Functional and phenotypic characterization of distinct porcine dendritic cells derived from periheral blood monocytes. Immunology. 2001;102:396–404. doi: 10.1046/j.1365-2567.2001.01200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan SSM, McConnell I, Blacklaws BA. Generation and characterization of ovine dendritic cells derived from peripheral blood monocytes. Immunology. 2002;107:366–72. doi: 10.1046/j.1365-2567.2002.01515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bienzle D, Reggeti F, Clark ME, Chow C. Immunophenotype and functional properties of feline dendritic cells derived drom blood and bone marrow. Vet Immunl Immunopathol. 2003;6:19–30. doi: 10.1016/s0165-2427(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 14.Mauel S, Steinbach F, Ludwig H. Monocyte-derived dendritic cells from horses differ from dendritic cells of humans and mice. Immunology. 2006;117:463–73. doi: 10.1111/j.1365-2567.2005.02319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang YS, Chi KH, Liao KW, Liu CC, Cheng CC, Lin YC, Cheng CH, Chu RM. Characterization of canine monocyte-derived dendritic cells with phenotypic and functional differentiation. Can J Vet Res. 2007;71:165–74. [PMC free article] [PubMed] [Google Scholar]
- 16.Dietze B, Cierpka E, Schafer M, Schill W, Lutz MB. An improved method to generate equine dendritic cells from peripheral blood mononuclear cells: divergent maturation programs by IL-4 and LPS. Immunobiology. 2008;213:751–8. doi: 10.1016/j.imbio.2008.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Cavatorta DJ, Erb HN, Flaminio MJ. Ex vivo generation of mature equine monocyte-derived dendritic cells. Vet Immunl Immunopathol. 2009;131:259–67. doi: 10.1016/j.vetimm.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 18.Sallusto F, Cella M, Danieli C, Lanzavecchia A. Denditic cells use maropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: downregulation by cytokines and bacterial products. J Exp Med. 1995;182:389–400. doi: 10.1084/jem.182.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou LJ, Tedder TF. Human blood dendriitc cells selectively express CD83, a member of the immunoglobulin superfamily. J Immunol. 1995;154:3821–35. [PubMed] [Google Scholar]
- 20.Mellman I, Turley SJ, Steinman RM. Antigen processing for amateurs and professionals. Trends Cell Biol. 1998;8:231–7. doi: 10.1016/s0962-8924(98)01276-8. [DOI] [PubMed] [Google Scholar]
- 21.Cochand L, Isler P, Songeon F, Nicod LP. Human lung dendritic cells have an immature phenotype with eifficient mannose receptors. Am J Respir Cell Mol Biol. 1999;21:547–54. doi: 10.1165/ajrcmb.21.5.3785. [DOI] [PubMed] [Google Scholar]
- 22.Wade CM, Giulotto E, Sigurdsson S, et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science. 2009;326:865–7. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins M, Ling V, Carreno BM. The B7 family of immune-regulatory ligands. Genome Biol. 2005;6:223. doi: 10.1186/gb-2005-6-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinbach F, Bischoff S, Freund H, Metzner-Flemisch S, Ibrahim S, Walter J, Wilke I, Mauel S. Clinical application of dendritic cells and interleukin-2 and tools to study activated T cells in horses – first results and implications for quality control. Vet Immunl Immunopathol. 2009;128:16–23. doi: 10.1016/j.vetimm.2008.10.317. [DOI] [PubMed] [Google Scholar]
- 25.Ibrahim S, Saunders K, Kydd JH, Lunn DP, Steinbach F. Screening of anti-human leukocyte monoclonal antibodies for reactivity with equine leukocytes. Vet Immunl Immunopathol. 2007;119:63–80. doi: 10.1016/j.vetimm.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 26.Schuler G. Murine epidermal Langerhans cells mature into potent immunostimulatory dendritic cells in vitro. J Exp Med. 1985;161:526–46. doi: 10.1084/jem.161.3.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang AL, Colmenero P, Purath U, et al. Natural killer cells trigger differentiation of monocytes into dendritic cells. Blood. 2007;110:2484–93. doi: 10.1182/blood-2007-02-076364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quah BJ, Warren HS, Parish CR. Monitoring lymphocyte proliferation in vitro and in vivo with the intracellular fluorescent dye carboxyfluorescein diacetate succinimidyl ester. Nat Protoc. 2007;2:2049–56. doi: 10.1038/nprot.2007.296. [DOI] [PubMed] [Google Scholar]
- 29.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–86. [DOI] [PubMed] [Google Scholar]
- 30.Cappelli K, Felicetti M, Capomaccio S, Spinsanti G, Silvestrelli M, Supplizi AV. Exercise induced stress in horses: selection of the most stable reference genes for quantitative RT-PCR normalization. BMC Mol Biol. 2008;9:49–56. doi: 10.1186/1471-2199-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.den Dekker E, Grefte S, Huijis T, et al. Monocyte cell surface gycosaminoglycans positively modulate IL-4-induced differentiation toward dendritic cells. J Immunol. 2008;180:3680–8. doi: 10.4049/jimmunol.180.6.3680. [DOI] [PubMed] [Google Scholar]
- 33.Wollenberg MM, Oppel T, Schottdorf E, Gunther S, Moderer M. Expression and function of the mannose receptor CD206 on epidermal dendritic cells in inflammatory skin diseases. J Invest Dermatol. 2002;118:327–34. doi: 10.1046/j.0022-202x.2001.01665.x. [DOI] [PubMed] [Google Scholar]
- 34.Klein E, Koch S, Borm B, Neumann J, Herzog V, Koch N, Bieber T. CD83 localization in a recycling compartment of immature human monocyte-derived dendritic cells. Int Immunol. 2005;17:477–87. doi: 10.1093/intimm/dxh228. [DOI] [PubMed] [Google Scholar]
- 35.Cao W, Lee SH, Lu J. CD83 is preformed inside monocytes, macrophages and dendritic cells, but it is only stably expressed on activated dendritic cells. Biochem J. 2005;385:85–93. doi: 10.1042/BJ20040741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jonuleit H, Kuhn U, Muller G, Steinbrink K, Paragnik L, Schmitt E, Knop J, Enk AH. Proinflammatory cytokines and prostagladins induce maturation of potent immunostimulatory dendriitc cells under fetal calf serum-free conditions. Eur J Immunol. 1997;27:3135–42. doi: 10.1002/eji.1830271209. [DOI] [PubMed] [Google Scholar]
- 37.Landi A, Babiuk LA, van Drunen Littel-van den Hurk S. Dendritic cells matured by a prostaglandin E2-containing cocktail can produce high levels of IL-12p70 and are more mature and Th1-biased than dendritic cells treated with TNF-α or LPS. Immunobiology. 2011;216:649–62. doi: 10.1016/j.imbio.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Sallusto F, Schaerli P, Loetscher P, Schaniel C, Lenig D, Mackay CR, Qin S, Lanzavecchia A. Rapid and coordinated switch in chemokine receptor expression during dendritic cell maturation. Eur J Immunol. 1998;28:2760–9. doi: 10.1002/(SICI)1521-4141(199809)28:09<2760::AID-IMMU2760>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 39.Sozzani S, Allavena P, D'Amico G, et al. Differential regulation of chemokine receptors during dendritic cell maturation: a model for their trafficking properties. J Immunol. 1998;161:1083–6. [PubMed] [Google Scholar]
- 40.Riol-Blanco L, Sanchez-Sanchez N, Torres A, Tejedor A, Narumiya S, Corbi AL, Sanchez-Mateos P, Rodriguez-Fernandez JL. The chemokine receptor CCR7 activates in dendriitc cells two signalling modules that independently regulate chemotaxis and migratory speed. J Immunol. 2005;174:4070–80. doi: 10.4049/jimmunol.174.7.4070. [DOI] [PubMed] [Google Scholar]
- 41.Berchtold S, Muhl-Zurbes P, Heufler C, Winklehner P, Schuler G, Steinkasserer A. Cloning, recombinant expression and biochemical characterization of the murine CD83 molecule which is specifically upregulated during dendritic cell maturation. FEBS Lett. 1999;461:211–6. doi: 10.1016/s0014-5793(99)01465-9. [DOI] [PubMed] [Google Scholar]
- 42.Kruse M, Rosorius O, Kratzer F, Bevec D, Kuhnt C, Steinkasserer A, Schuler G, Hauber J. Inhibition of CD83 cells surface expression during dendritic cell maturation by interference with nuclear export of CD83 mRNA. J Expl Med. 2000;191:1581–9. doi: 10.1084/jem.191.9.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuwano Y, Prazma CM, Yazawa N, et al. CD83 influences cell-surface MHC class II expression on B cells and other antigen-presenting cells. Int Immunol. 2007;19:977–92. doi: 10.1093/intimm/dxm067. [DOI] [PubMed] [Google Scholar]
- 44.Tze LE, Horikawa K, Domaschenz H, et al. CD83 increases MHC II and CD86 on dendritic cells by opposing IL-10-driven MARCH1-mediated ubiquitination and degradation. J Expl Med. 2011;208:149–65. doi: 10.1084/jem.20092203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iborra S, Izquierdo H, Martínez-López M, Blanco-Menéndez N, Reis e Sousa C, Sancho D. The DC receptor DNGR-1 mediates cross-priming of CTLs during vaccinia virus infection in mice. J Clin Invest. 2012;122:1628–43. doi: 10.1172/JCI60660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zelenay S, Keller AM, Whitney PG, et al. The dendritic cell receptor DNGR-1 controls endocytic handling of necrotic cell antigens to favor cross-priming of CTLs in virus-infected mice. J Clin Invest. 2012;122:1615–27. doi: 10.1172/JCI60644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nestle FO, Farkas A, Conrad C. Dendritic-cell-based therapeutic vaccination against cancer. Curr Opin Immunol. 2005;17:163–9. doi: 10.1016/j.coi.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 48.Oelke M, Maus MV, Didiano D, June CH, Mackensen A, Schneck JP. Ex vivo induction and expansion of antigen-specific cytotoxic T cells by HLA-Ig-coated artificial antigen-presenting cells. Nat Med. 2003;9:619–24. doi: 10.1038/nm869. [DOI] [PubMed] [Google Scholar]
- 49.Ponnazhagan S, Mahendra G, Curiel DT, Shaw DR. Adeno-associated virus type 2-mediated transduction of human monocyte-derived dendritic cells: implications for ex vivo immunotherapy. J Virol. 2001;75:9493–501. doi: 10.1128/JVI.75.19.9493-9501.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuler G, Schuler-Thurner B, Steinman RM. The use of dendritic cells in cancer immunotherapy. Curr Opin Immunol. 2003;15:138–47. doi: 10.1016/s0952-7915(03)00015-3. [DOI] [PubMed] [Google Scholar]
- 51.Brown JA, Dorfman DM, Ma F, Sullivan EL, Munoz O, Wood CR, Greenfield EA, Freeman GJ. Blockade of programmed death-1 ligands on dendritic cells enhances T cell activation and cytokine production. J Immunol. 2003;170:1257–66. doi: 10.4049/jimmunol.170.3.1257. [DOI] [PubMed] [Google Scholar]
- 52.Carter LL, Fouser LA, Jussif J, et al. PD-1:PD-L inhibitory pathway affects both CD4+ and CD8+ T cells and is overcome by IL-2. Eur J Immunol. 2002;32:634–43. doi: 10.1002/1521-4141(200203)32:3<634::AID-IMMU634>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 53.Curiel TJ, Wei S, Dong H, et al. Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity. Nat Med. 2003;9:562–7. doi: 10.1038/nm863. [DOI] [PubMed] [Google Scholar]
- 54.Freeman GJ, Long AJ, Iwai Y, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–34. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Latchman Y, Wood CR, Chernova T. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2:261–8. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 56.Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family costimulates T-cell proliferation and interleukin-10 secretion. Nat Med. 1999;5:1365–9. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 57.Tseng S, Otsujim M, Gorski K, et al. B7-DC, a new dendritic cell molecule with potent costimulatory properties for T cells. J Exp Med. 2001;193:839–45. doi: 10.1084/jem.193.7.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aicher A, Hayden-Ledbetter M, Brady WA, et al. Characterization of human inducible costimulator ligand expression and function. J Immunol. 2000;164:4689–96. doi: 10.4049/jimmunol.164.9.4689. [DOI] [PubMed] [Google Scholar]
- 59.Hutloff A, Dittrich AM, Beier KC, Eljaschewitsch B, Kraft R, Anagnostopoulos I, Kroczek RA. ICOS is an inducible T-cell co-stimulator structurally and functionally related to CD28. Nature. 1999;387:263–6. doi: 10.1038/16717. [DOI] [PubMed] [Google Scholar]
- 60.Sperling AI, Bluestone JA. ICOS costimulation: it's not just for TH2 cells anymore. Nature. 2001;2:573–4. doi: 10.1038/89709. [DOI] [PubMed] [Google Scholar]
- 61.Zhang GB, Zhou H, Chen YJ, et al. Characterization and application of two novel monoclonal antibodies against 2IgB7-H3: expression analysis of 2IgB7-H3 on dendritic cells and tumor cells. Tissue Antigens. 2005;66:83–92. doi: 10.1111/j.1399-0039.2005.00449.x. [DOI] [PubMed] [Google Scholar]
- 62.Chapoval AI, Ni J, Lau JS, et al. B7-H3: a costimulatory molecule for T cell activation and IFN-γ production. Nature. 2001;2:269–74. doi: 10.1038/85339. [DOI] [PubMed] [Google Scholar]
- 63.Wang S, Zhu G, Chapoval A, Dong H, Tamada K, Ni J, Chen L. Costimulation of T cells by B7-H2 a B7-like molecule that binds ICOS. Blood. 2000;96:2808–13. [PubMed] [Google Scholar]
- 64.Ling V, Wua PW, Spauldinga V, Kieleczawaa J, Luxenberga D, Carrenoa BM, Collinsa M. Duplication of primate and rodent B7-H3 immunoglobulin V- and C-like domains: divergent history of functional redundancy and exon loss. Genomics. 2003;82:365–77. doi: 10.1016/s0888-7543(03)00126-5. [DOI] [PubMed] [Google Scholar]
- 65.Prasad DVR, Nguyen T, Li Z, Yang Y, Duong J, Wang Y, Dong C. Murine B7-H3 is a negative regulator of T cells. J Immunol. 2004;173:2500–6. doi: 10.4049/jimmunol.173.4.2500. [DOI] [PubMed] [Google Scholar]
- 66.Suh WK, Gajewska BU, Okada H, et al. The B7 family member B7-H3 preferentially down-regulates T helper type 1-mediated immune responses. Nat Immunol. 2003;4:899–906. doi: 10.1038/ni967. [DOI] [PubMed] [Google Scholar]
- 67.Allavena P, Sica A, Vecchi A, Locati M, Sozzani S, Mantovani A. The chemokine receptor switch paradigm and dendritic cell migration: its significance in tumor tissues. Immunol Rev. 2000;177:141–9. doi: 10.1034/j.1600-065x.2000.17714.x. [DOI] [PubMed] [Google Scholar]
- 68.Dieu M, Vanbervliet B, Vicari A, et al. Selective recruitment of immature and mature dendritic cells by distinct chemokines expressed in different anatomic sites. J Exp Med. 1998;188:373–86. doi: 10.1084/jem.188.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yanagihara S, Komura E, Nagafune J, Watarai H, Yamaguchi Y. EBI1/CCR7 Is a new member of dendritic cell chemokine receptor that is up-regulated upon maturation. J Immunol. 1998;161:3096–102. [PubMed] [Google Scholar]
- 70.Cravens PD, Lipsky PE. Dendritic cells, chemokine receptors and autoimmune inflammatory diseases. Immunol Cell Biol. 2002;80:497–505. doi: 10.1046/j.1440-1711.2002.01118.x. [DOI] [PubMed] [Google Scholar]
- 71.Song A, Nikolcheva T, Krensky AM. Transcriptional regulation of RNATES expression in T lymphocytes. Immunol Rev. 2000;177:236–45. doi: 10.1034/j.1600-065x.2000.17610.x. [DOI] [PubMed] [Google Scholar]
- 72.D'Ambrosio D, Iellem A, Bonecchi R, Mazzeo D, Sozzani S, Mantovani A, Sinigaglia F. Cutting edge: selective up-regulation of chemokine receptors CCR4 and CCR8 upon activation of polarized human type 2 Th cells. J Immunol. 1998;161:5111–5. [PubMed] [Google Scholar]
- 73.Imai T, Nagira M, Takagi S, et al. Selective recruitment of CCR4-bearing TH2 cells toward antigen-presenting cells by the CC chemokines thymus and activation-regulated chemokine and macrophage-derived chemokine. Int Immunol. 1999;11:81–8. doi: 10.1093/intimm/11.1.81. [DOI] [PubMed] [Google Scholar]
- 74.Vissers JLM, Hartgers FC, Lindhout E, Flgdor CG, Adema GJ. BLC (CXCL13) is expressed by different dendritic cell subsets in vitro and in vivo. Eur J Immunol. 2001;31:1544–9. doi: 10.1002/1521-4141(200105)31:5<1544::AID-IMMU1544>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 75.Chensue SW, Warmington KS, Ruth JH, Sanghi PS, Lincoln P, Kunkel SL. Role of monocyte chemoattractant protein-1 (MCP-1) in Th1 (mycobacterial) and Th2 (schistosomal) antigen-induced granuloma formation. J Immunol. 1996;157:4602–8. [PubMed] [Google Scholar]
- 76.Gu L, Tseng S, Horner RM, Tam C, Loda M, Rollins J. Control of TH2 polarization by the chemokine monocyte chemoattractant protein-1. Nature. 2000;404:407–11. doi: 10.1038/35006097. [DOI] [PubMed] [Google Scholar]
- 77.Padovan E, Spagnoli GC, Ferrantini M, Heberer M. IFN-α2a induces IP-10/CXCL10 and MIG/CXCL9 production in monocyte-derived dendritic cells and enhances their capacity to attract and stimulate CD8+ effector T cells. J Leukoc Biol. 2002;71:669–76. [PubMed] [Google Scholar]
- 78.Guzylack-Piriou L, Piersma S, McCullough K, Summerfield A. Role of natural interferon-producing cells and T lymphocytes in porcine monocyte-derived dendritic cell maturation. Immunology. 2006;118:78–87. doi: 10.1111/j.1365-2567.2006.02343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ricklin Gutzwiller ME, Moulin HR, Zurbriggen A, Roosje P, Summerfield A. Comparative analysis of canine monocyte- and bone-marrow-derived dendritic cells. Vet Res. 2010;41:40–51. doi: 10.1051/vetres/2010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guilliams M, Henri S, Tamoutounour S, Ardouin L, Schwartz-Cornil I, Dalod M, Malissen B. From skin dendritic cells to a simplified classification of human and mouse dendritic cell subsets. Eur J Immunol. 2010;40:2089–94. doi: 10.1002/eji.201040498. [DOI] [PubMed] [Google Scholar]


