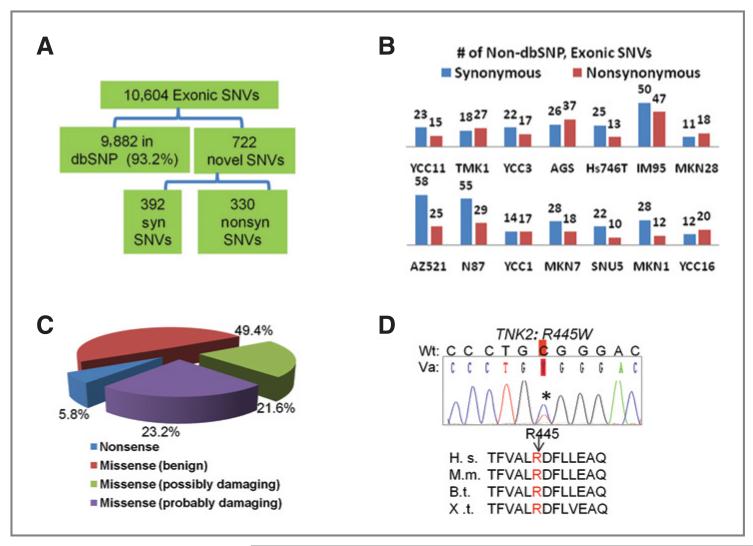
Figure 2.
Identification of kinase microgenomic variants. A, total number of variants identified in the cell lines and their breakdown across various categories (dbSNP or novel, synonymous, syn, or nonsynonymous). B, number of novel exonic synonymous and nonsynonymous SNVs in individual cell lines. C, PolyPhen in silico assessment of functional impact of novel exonic nonsynonymous SNVs identified (see the text). D, sequencing chromatograph of a germline variant TNK2(R445W) found in 1 of 48 GC patients and in 2 cell lines (MKN28 and IM95). R445 locates in the kinase domain of TNK2 and is conserved across multiple species. H.s., Homo sapiens; M. m., Mus musculus; B.t., Bos Taurus; X.t., Xenopus tropicalis. Wt, wild-type; Va, variant.