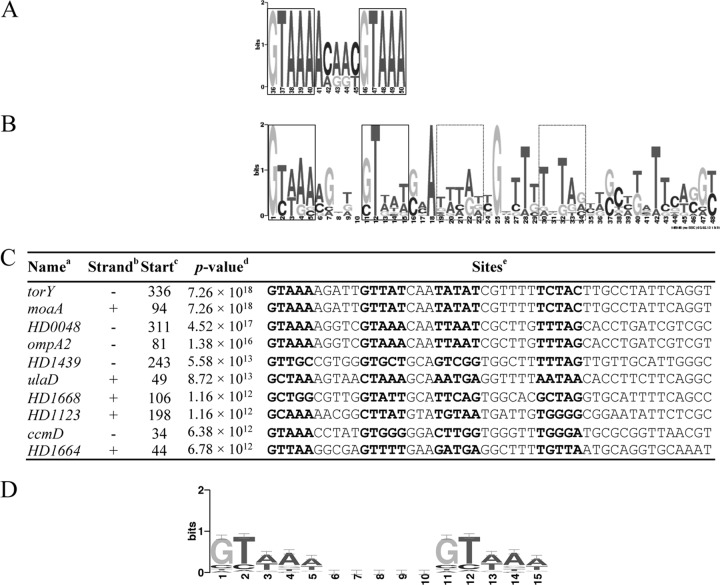
Fig 4.
The H. ducreyi and E. coli CpxR binding motifs. (A) Sequence logo of the E. coli CpxR binding sites. The sequence logo was generated with the 450-bp promoter regions from 10 known Cpx target operons (12). The core regions in the motif are boxed. (B) The extended CpxR binding motif in H. ducreyi. The extended logo was generated by the MEME algorithm with the 450-bp upstream promoter sequences from the 66 downregulated targets identified in the 35000HPΔcpxA-versus-35000HPΔcpxR comparison in the stationary phase. The putative core regions in the extended motif are boxed. (C) Multiple-sequence alignment of the extended CpxR binding motif in the 10 promoter regions that contained the most significant matches. Superscript letters: a, genes in which the motif was identified; b, location of the extended motif on the template strand (+) or the complementary strand (−); c, start position of the binding site relative to the ATG start codon; d, P value for the extended motif; e, sequence of the predicted motif with the putative core regions in bold. (D) Sequence logo of H. ducreyi CpxR recognition weight matrix. The matrix frequency of a single GTAAA repeat is obtained by averaging the two parts of the first tandem repeat shown in Fig. 4B. The same nucleotide frequency was used for both GTAAA repeats. The 5-bp linker was assigned with a background frequency that was given by the MEME algorithm.