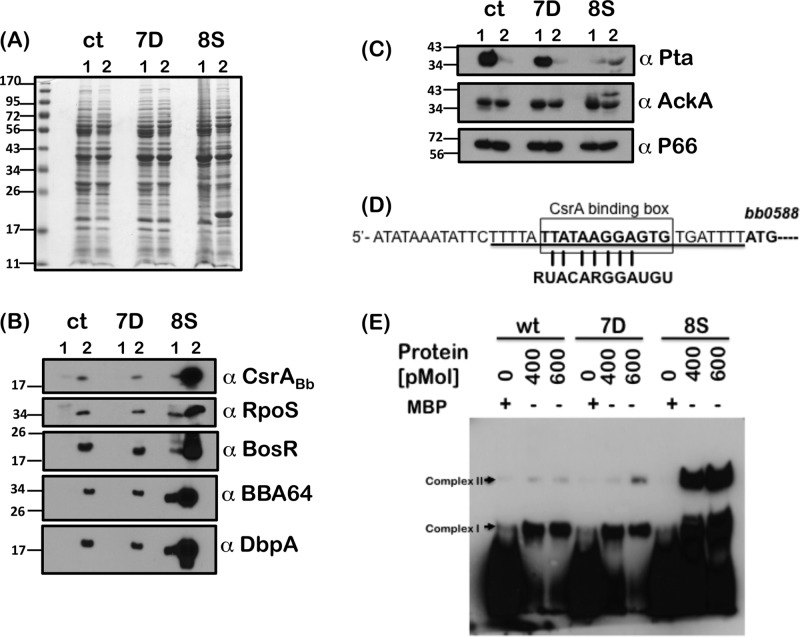
Fig 6.
Phenotypic analysis of csrABb mutants with site-specific alterations or C-terminal deletion. (A) Proteins from cis-complemented strains carrying wild-type CsrABb (ct), CsrABb with a 7-amino-acid C-terminal deletion (7D), or CsrABb with substitutions of 8 critical residues (8S) were propagated under conditions mimicking the tick vector conditions before (23°C/pH 7.6) or after (37°C/pH 6.8) a blood meal. Proteins were separated on an SDS–12.5% PAGE and stained with Coomassie blue. (B) Immunoblot analysis performed with antisera against CsrABb, RpoS, BosR, BBA64, and DbpA. (C) Immunoblot analysis of ct, 7D, and 8S strains with antisera against Pta, AckA, and P66 (loading control). Molecular masses in kilodaltons are indicated to the left. (D) The CsrABb binding box of the 5′UTR of bb0588 to bb0589 known to regulate levels of Pta was used to generate a biotinylated RNA probe (26). (E) Efficiency of binding of recombinant wt and mutant CsrABb proteins to the 5′UTR of bb0588 to bb0589. A biotinylated RNA probe (20 fmol) was incubated with purified recombinant wt, 7D, and 8S fused to maltose binding protein (MBP) at the concentrations indicated above each lane. MBP alone was used at a 600-pmol concentration as a control (MBP lanes). Blots were developed using enhanced chemiluminescence (ECL) as described in Materials and Methods and represent one of three independent experiments.