Fig 1.
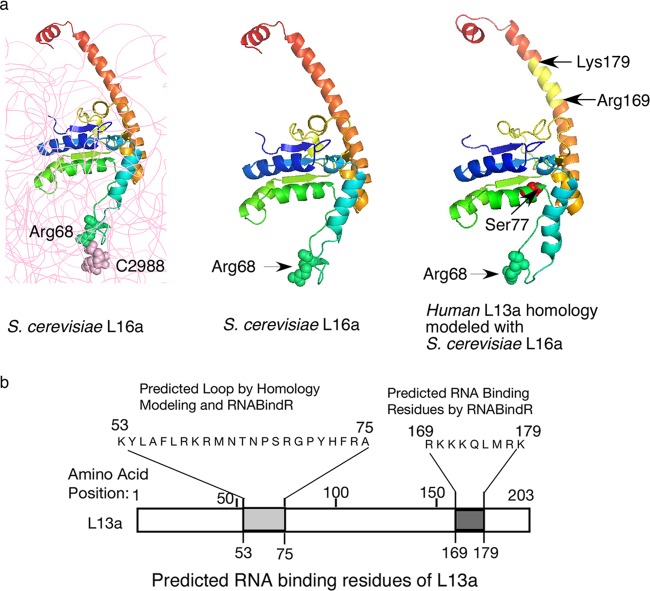
L13a protein structure and the putative rRNA binding residues. (a, left and middle) The structure of yeast S. cerevisiae L16a (L13a homologue) depicted as ribbon diagrams based on the crystal structure (Protein Data Bank codes 3U5E and 3O58). In the left panel, rRNA is shown in gray; L16a Arg68 forms van der Waals interactions with 25S rRNA C2988. In the right panel, the predicted structural model of human L13a, generated by using the S. cerevisiae L16a structure as a template for comparative modeling, is shown. van der Waals radii of Arg68 and C2988 are shown. The IFN-γ-mediated phosphorylation site at Ser77 and the RNA binding residues (Arg169 to Lys179) predicted by RNABindR are shown. (b) Potential L13a RNA binding regions predicted by homology modeling using the S. cerevisiae L16a structure and/or the Web-based RNABindR server (http://einstein.cs.iastate.edu/RNABindR/).