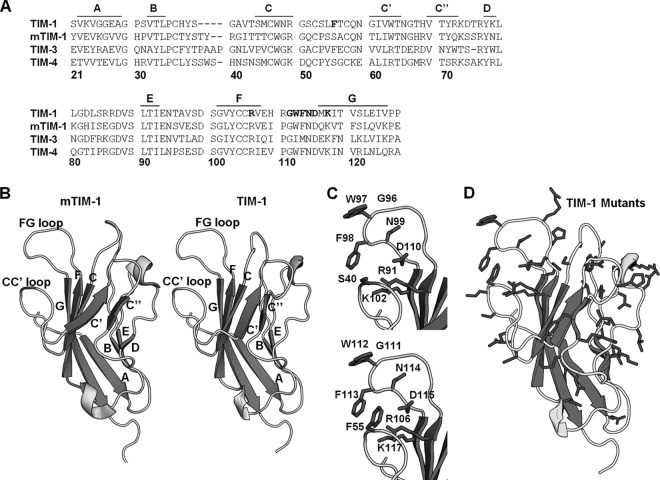
Fig 1.
TIM-1 threaded on mTIM-1 crystal structure. (A) Amino acid sequences of TIM-1, mTIM-1, TIM-3, and TIM-4 IgV domains aligned using the Clustal W software program. Residues in the TIM-1 PtdSer binding pocket that were mutated are shown in bold. β-Sheets are marked above the alignment with lines and corresponding letters. Residue numbering is based on the TIM-1 sequence. (B) Structures of mTIM-1 IgV (20R8) (left) and a threaded model of TIM-1 IgV (right). β-Sheets are labeled with their corresponding letters, assigned by Santiago et al. (40). (C) PtdSer binding residues of mTIM-1 (above) and TIM-1 (below) are shown on each structure. (D) All residues in the TIM-1 IgV domain mutated in this study are highlighted in black.