Abstract
Eighty percent of malignant tumors that develop in the central nervous system are malignant gliomas, which are essentially incurable. Here, we discuss how recent sequencing studies are identifying unexpected drivers of gliomagenesis, including mutations in isocitrate dehydrogenase 1 and the NF-κB pathway, and how genome-wide analyses are reshaping the classification schemes for tumors and enhancing prognostic value of molecular markers. We discuss the controversies surrounding glioma stem cells and explore how the integration of new molecular data allows for the generation of more informative animal models to advance our knowledge of glioma's origin, progression, and treatment.
Although the total incidence of primary central nervous system (CNS) tumors is only about 18.7 per 100,000 persons in the United States, 80% of the malignant tumors in the CNS are malignant gliomas, which are essentially incurable (CBTRUS, 2009). Gliomas display histological similarities to glial cells, including astrocytes and oligodendrocytes. According to the 2007 World Health Organization (WHO) classification, malignant gliomas can be classified according to which cell they most resemble, such as astrocytomas, oligodendrogliomas, or oligoastrocytomas. More than half of gliomas are glioblastoma multiforme (GBM, grade IV astrocytoma), one of the most aggressive cancers (Louis et al., 2007). Despite decades of concerted effort and advances in surgery, radiation, and chemotherapy, the overall 5 year survival rate of GBM remains less than 5% and is even worse for elderly patients (CBTRUS, 2009). This dismal clinical outcome makes glioma an urgent subject of cancer research. Here, we discuss current advances in genomic analysis and genetic modeling of glioma and how these developments influence strategies for therapeutic intervention in this deadly disease.
Genetics of Glioma
Glioma Core Signaling Pathways
In the past two decades, cytogenetic and molecular genetic studies have identified a number of recurrent chromosomal abnormalities and genetic alterations in malignant gliomas, particularly in GBM. Advances in molecular technologies, especially high-density microarray and genome sequencing, have made it possible to evaluate genetic and epigenetic changes in these tumors at the genome-wide level. In a comprehensive study carried out by The Cancer Genome Atlas (TCGA) project, 601 cancer-related candidate genes were sequenced in more than 200 human GBM samples (TCGA, 2008). The project also analyzed genome-wide DNA copy number changes, DNA methylation status, and protein-coding and noncoding RNA expression (TCGA, 2008). A similar but complementary study by Parsons et al. sequenced 20,661 protein-encoding genes in 22 GBM samples and integrated the genetic alteration information with DNA copy number and gene expression profiles (Parsons et al., 2008).
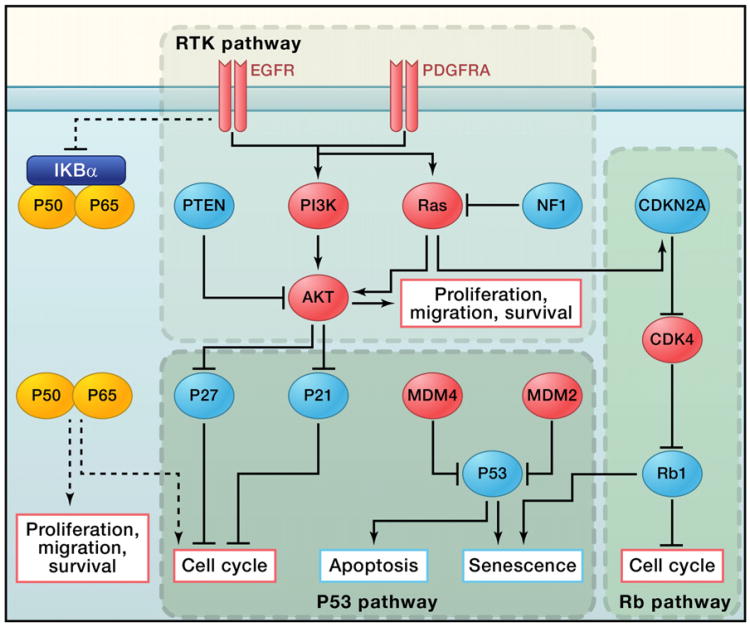
These integrative genomic studies provided a comprehensive view of the complicated genomic landscape of GBM, revealing a set of core signaling pathways commonly activated in GBM (Figure 1): the P53 pathway, the RB pathway, and the RTK pathway (TCGA, 2008; Parsons et al., 2008). Furnari et al. have written a detailed review of these pathways in glioma (Furnari et al., 2007). The majority of GBM tumors have genetic alterations in all three pathways, which helps to fuel cell proliferation and enhance cell survival while allowing the tumor cell to escape from cell-cycle checkpoints, senescence, and apoptosis. In addition to confirming known genetic events, the TCGA sequencing data also provided somatic mutation information at the base pair level, revealing potential new roles for known tumor suppressors/oncogenes in GBM as well as new cancer driver genes. For example, it has long been observed that patients with germline mutations in the tumor suppressor gene responsible for neurofibromatosis type 1 (NF1) have an increased incidence of malignant glioma (Gutmann et al., 2002; Friedman, 1999). Studies in genetic mouse models have also strongly suggested a causal role for NF1 mutation in glioma tumorigenesis (Alcantara Llaguno et al., 2009; Kwon et al., 2008; Zhu et al., 2005). However, the involvement of NF1 mutation in sporadic human GBM remained underappreciated until the TCGA project reported that 47 of the 206 patient samples, or 23%, had NF1 mutations or deletions, ranking it as the third most frequently somatically mutated gene among the 601 genes sequenced (TCGA, 2008).
Figure 1. Core Signaling Pathways in Glioma Tumorigenesis.
The receptor tyrosine kinase (RTK), p53, and Rb pathways are the core signaling pathways in glioma oncogenesis. Red indicates oncogenes that are either overexpressed or amplified in GBM samples, and blue indicates tumor suppressor genes that are somatically mutated or deleted (except for P27 and P21).
In addition to the core signaling pathways identified through genome-wide screening studies, Harsh et al. recently reported that heterozygous deletion of the NF-κB inhibitor α (NFKBIA) gene was present in a quarter of GBM samples (Bredel et al., 2011). The NFKBIA gene encodes the protein IκBα, a crucial negative regulator in the canonical NF-κB signaling pathway. Under basal conditions, IκBα sequesters the NF-κB transcription factor heterodimer (p50/p65) in the cytoplasm. Upon stimulation with ligand such as tumor necrosis factor α (TNF-α) or lipopolysaccharide (LPS), IκBα is phosphorylated by the signalosome (Karin, 2006). This phosphorylation leads to rapid ubiquitination and degradation of IκBα, which releases the inhibition of NF-κB and allows translocation of p50/p65 into the nucleus to activate transcription of downstream target genes, including many cytokines that can promote tumor growth and infiltration (Karin, 2006).
In GBM, NFKBIA deletion and EGFR amplification are mutually exclusive, raising the possibility that the two genetic events converge on the same pathway. Indeed, overexpression of NFKBIA reduced the viability of primary glioma cells in which NFKBIA was downregulated or EGFR was upregulated (Bredel et al., 2011). In addition, both genetic events were associated with similar prognostic outcome, which is inferior to that of patients with normal expression levels of these two genes. However, the detailed molecular mechanism for the role of NF-κB in glioma development and progression and its connection with EGFR signaling remain to be investigated.
IDH Mutations in Glioma
Among the various genomic efforts to characterize gliomas, the biggest surprise came from the genome-wide exon sequencing project, in which R132 mutations of isocitrate dehydrogenase 1 (IDH1) were observed in 12% of the GBM samples (Parsons et al., 2008). Subsequent studies revealed that as many as 70%–90%of the grade II/III gliomas harbored this IDH1 mutation (Yan et al., 2009). Other studies have demonstrated that some gliomas contain IDH2 R172 mutations, an analog to IDH1 R132, at a lower frequency (Yan et al., 2009). Two additional point mutations, IDH1 R100 and IDH2 R140, have been reported in AML but have not been observed in human glioma samples (Green and Beer, 2010).
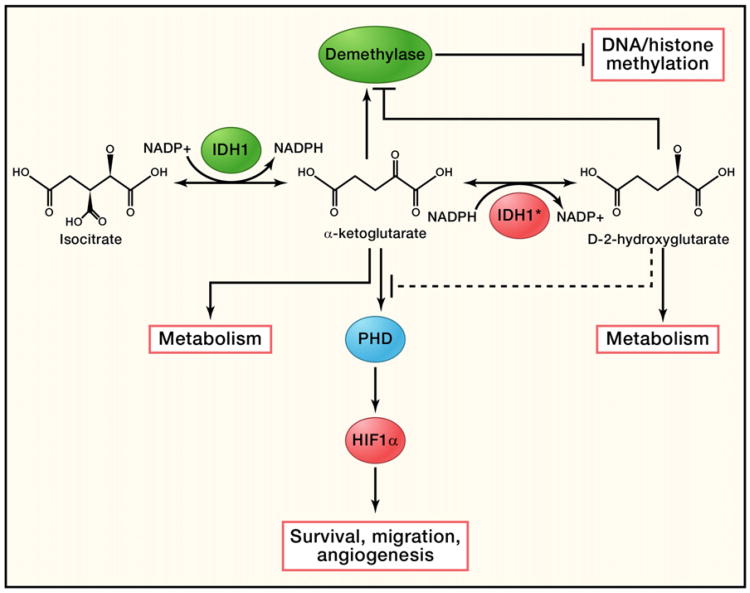
Normally, IDH1 and IDH2 convert isocitrate to α-ketoglutarate (α-KG) while at the same time reducing NADP+ to NADPH in the cytosol and mitochondria, respectively (Figure 2) (Reitman et al., 2010). It was initially hypothesized that the IDH1 R132 and IDH2 R172 mutations reduced the enzyme's ability to generate α-KG (Yan et al., 2009). Subsequent studies reported a dominant-negative role for IDH1 R132 and suggested a tumor suppressor function for IDH1/2 (Zhao et al., 2009). However, human genetic data showed that the mutations were always observed on a specific residue and only in one allele of the gene. These apparently narrow constraints on the nature of the mutations raised the possibility of neomorphic (gain-of-function) mutations (Green and Beer, 2010; Reitman et al., 2010; Yan et al., 2009). Follow-up studies discovered that the IDH1/2 mutations had an NADPH-dependent ability to convert α-KG to D-2-hydroxyglutarate (D-2HG) (Figure 2), supporting a pro-oncogenic role for IDH1/2 (Dang et al., 2010; Ward et al., 2010). Consistent with this model, knocking down wild-type IDH1 in a glioma cell line slowed cell growth, and levels of 2HG were found to be 10-fold higher in IDH1/2 mutated glioma or leukemia samples (Dang et al., 2010; Ward et al., 2010).
Figure 2. Function of Normal and Mutated IDH1.
Wild-type IDH1 catalyzes isocitrate to form α-ketoglutarate and convert NADP+ to NADPH at the same time. A mutated form of IDH1 can convert α-ketoglutarate to D2-hydroxyglutarate in an NADPH-dependent manner. Excessive D2-hy-droxyglutarate results in cellular stress as well as metabolic changes. It could also potentially act as a competitive substrate to inhibit DNA/histone methyltransferases and prolyl hydroxylases (PHDs), resulting in DNA/histone hypomethylation or activation of HIF-1α, which can be further accelerated by the lack of α-ketoglutarate, as α-ketoglutarate is a key substrate for both PHDs and DNA/histone methyltransferases.
Despite this intriguing data, the mechanism by which IDH1/2 mutations transform cells is far from clear. The discovery of a neomorphic enzymatic function for IDH1/2 raises the possibility that D-2HG may act as an oncometabolite (Dang et al., 2010; Ward et al., 2010). It has been reported that increased levels of D-2HG in cells caused oxidative stress in rat brains (Latini et al., 2003), which could potentially promote oncogenesis. In the clinic, high levels of 2HG have been linked to a rare neurometabolic disorder called D-2-hydroxyglutaric aciduria (Kranendijk et al., 2010). A subset of patients with this disease was found to harbor IDH2 R140 mutations. Though the patients showed significantly higher levels of D-2HG compared to leukemia or glioma patients with IDH1/2 mutations, they did not develop gliomas, leukemia, or other malignancies (Kranendijk et al., 2010). In glioma patients, IDH1/2 mutations usually coexist with TP53 mutations (Yan et al., 2009). It is possible that, like other oncogenic stresses that trigger cell death and senescence, cells with extremely high levels of 2HG may be restrained from further malignant transformation by a similar checkpoint mechanism.
In addition, other substrates/products affected by the IDH1/2 mutations may also contribute to oncogenesis. Disrupting the balance of NADP+/NADPH is likely to result in a broad spectrum of cellular reactions, and α-KG is a key component of multiple pathways. For example, α-KG is a key substrate for prolyl hydroxylase domain proteins (PHDs) to catalyze hydroxylation of hypoxia-inducible factor (HIF), a key regulator of angiogenesis (Figure 2) (Semenza, 2010). This hydroxylation allows HIF to be recognized and targeted for ubiquitin-mediated protein degradation. It has been observed that overexpression of IDH1 R132 in glioma cell lines resulted in increased levels of HIF-1α (Zhao et al., 2009). However, whether cells with IDH1/2 mutations have lower levels of α-KG is still controversial (Dang et al., 2010; Zhao et al., 2009). Given the structural similarity of α-KG and D-2HG, it is also possible that the new product D-2HG can compete with α-KG. Such competition has been linked to the oncogenic mechanism of succinate dehydrogenase (SDH) and fumarate hydratase (FH) mutations, in which accumulated succinate and fumarate compete with α-KG to inhibit the activity of PHDs (Semenza, 2010). In addition, α-KG is a substrate for particular histone and DNA demethylation enzymes (Figure 2). Reducing α-KG levels or levels of competing substrate would likely affect global gene expression. Indeed, 2-HG can inhibit multiple α-KG-dependent dioxygenases, which are important for DNA/histone demethylation (Xu et al., 2011). Consistent with this idea, gliomas with IDH1 mutations showed significantly higher frequency of the CpG island methylator (CIM) phenotype and increased histone demethylation (Noushmehr et al., 2010).
Molecular Subclassification of Gliomas
Subclassification of GBM by congruence of genomic features has taken precedence in the field. A detailed summary of the recent progress and problems related to this topic can be found in a recent review by Vitucci et al. (Vitucci et al., 2011). In general, genome-wide studies have revealed that tumor histology correlates with distinct gene expression signatures. Furthermore, molecular profiles can identify subclasses of tumors that would otherwise be indistinguishable by standard morphological methods. One such example is primary and secondary GBM. Although the histology of both types of GBM is identical, primary GBM is thought to arise de novo, and secondary GBM has a longer history of disease progression from lower-grade tumors (Ohgaki and Kleihues, 2007). Recently, however, several groups have used genome-wide analyses to successfully categorize these two subtypes based on their gene expression profiles (Maher et al., 2006; Tso et al., 2006).
Depending on the sample pool and analysis methods, different studies have reported different numbers of subclasses. For example, Li et al. published their molecular analysis of glioma using two different unsupervised methods; they reported two main types, oligodendroglioma-rich (O) and glioblastoma-rich (G), which could be further divided into six subtypes (Li et al., 2009a). Verhaak et al. performed unsupervised clustering analysis of the TCGA GBM data set and grouped the tumors into four subtypes termed proneural (PN), neural (NL), mesenchymal (MES), and classical (CL) (Verhaak et al., 2010). The PN and MES subtypes shared significant overlap with previous studies. Integration of genetic alteration events revealed that PN, MES, and CL subtypes were associated with aberrant PDGFRA/IDH1, NF1, and EGFR status, respectively. Gravendeel et al. also used an unsupervised algorithm to classify profiles of 276 gliomas into 24 different molecular “clusters,” or subtypes (Gravendeel et al., 2009). A similar proneural subtype (C17) was also identified. A large number of these classification studies have now been carried out and provide interesting insights into the molecular nature of these tumors, as well additional questions and problems to pursue (Vitucci et al., 2011).
The prognostic value of the molecular subclassification has also been evaluated, with several studies suggesting that gliomas with expression of genes associated with neurogenesis (proneural subtype) generally correlate with marginally improved survival (Vitucci et al., 2011). In contrast, gliomas with mesenchymal gene expression (mesenchymal subtype) usually have a poorer outcome (1 year for proneural versus 0.6 years for mesenchymal) (Vitucci et al., 2011). In an early study by Phillips et al., the proneural subtype included a significant proportion of grade III gliomas, which have a more favorable outcome compared to the more aggressive GBM (Phillips et al., 2006). Thus, it is possible that the difference in patient survival merely reflects the known survival differences associated with tumor grade. This problem was emphasized by the TCGA study in which only GBM samples were analyzed. No prognostic differences were observed among the four different subtypes revealed in the TCGA study (Verhaak et al., 2010). However, the C17 (PN) subtype in the Gravendeel study was found to have prolonged survival in either all of the gliomas or in the pure GBM population, confirming that the previous prognostic value of the molecular markers was not just the consequence of the selection methods, where markers were chosen based on their association with outcome in the training data sets (Gravendeel et al., 2009).
Although gliomas with proliferative or mesenchymal characteristics generally have a worse outcome, several studies have confirmed that these subtypes are also more sensitive to combinational radiation and chemotherapy, which could reflect the higher proliferative index or higher levels of microvascular endothelial proliferation (Gravendeel et al., 2009; Verhaak et al., 2010). Clinical trials designed based on this molecular subtype information are ongoing. At the same time, molecular mechanisms underlying the mesenchymal transformation of glioma have been studied by sophisticated in silico network analysis based on gene expression data and subtype classification. These data point to overexpression of transcription factor C/EBPβ and activation of STAT3 as driving forces of mesenchymal transformation (Carro et al., 2010).
Lessons from Genomic Studies
The wealth of genomic data that is now available for glioma provides tremendous opportunity to impact both basic research and clinical outcomes. The identification of previously unknown genetic alterations (e.g., IDH1/2, NF1, ERBB2, NFKBIA) provides opportunities for drug development against new therapeutic targets. Currently, genetic information is becoming more useful in making a clinical diagnosis and formulating treatment plans. Though decisions regarding glioma treatment are still mainly based on traditional pathology relying on histology, molecular genetic assessment now has an increasing role (Jansen et al., 2010). One such successful example in glioma is 1p/19q loss, which frequently occurs in oligodendrogliomas (Jansen et al., 2010). Patients with high-grade oligodendrogliomas that also harbor these genetic events have a better prognosis. In low-grade oligodendrogliomas, 1p/19q loss further dictates favorable progression-free survival after temozolomide treatment (Jansen et al., 2010). It has also been reported that coexpression of EGFRvIII and the tumor suppressor Pten associates with clinical response to EGFR kinase inhibitor in a recent clinical trial (Mellinghoff et al., 2005).
The discovery of core signaling pathways in glioma and the observation that several components of the same pathways are subject to mutagenesis in tumors also suggested a pathway targeting strategy, rather than targeting a single gene, for future drug discovery efforts (TCGA, 2008; Parsons et al., 2008). Consistent with this concept, coactivation of several tyrosine kinases was observed in both xenograft and primary human GBM specimens. In a cell culture system, targeting these RTKs concurrently using combinations of inhibitors and/or RNAi showed significantly better response than treating with a single agent alone (Stommel et al., 2007).
Continued accumulation of molecular information about glioma has also led to a reshaping of the classification scheme. For example, it was initially discovered that lower-grade gliomas have a higher frequency of TP53 mutation and PDGFRA expression, thus classifying these two events as markers of secondary GBM (Ohgaki and Kleihues, 2007). However, TP53 was also found to be the most frequent somatically mutated gene by the TCGA project, which predominantly studied primary GBM samples (TCGA, 2008). Likewise, in the same data set, a subset of GBMs was also found to overexpress PDGFRA (TCGA, 2008; Verhaak et al., 2010). These findings, on the one hand, demonstrate the need to identify additional markers for secondary GBM. Examples of additional markers are the recently identified IDH1/2 mutations, which have mainly been found in grade II-III gliomas (Parsons et al., 2008; Yan et al., 2009). On the other hand, it is conceivable that primary GBMs actually develop long before the patient becomes symptomatic. Consistent with this idea and previous mouse genetic studies (Zhu et al., 2005), bioinformatic efforts to reconstruct the temporal sequence of mutations in GBM using the TCGA data set indicated that the TP53 gene is likely mutated first in the tumorigenic process (Attolini et al., 2010). It is thus very desirable to develop diagnostic techniques to identify those early events for therapeutic intervention because patients with lower-grade gliomas have a significantly better prognosis (Ohgaki and Kleihues, 2007).
Genetic Mouse Models of Glioma
The laboratory mouse shares extensive molecular and physiological similarities to humans and is a powerful tool for studying cancer. Unlike invertebrate model systems, tumor development in mice is accompanied by other complex processes such as angiogenesis and metastasis, similar to those in human cancer. More importantly, mouse tumor models provide temporally and genetically controlled systems for studying the tumorigenic process as well as response to treatment. However, initial efforts to create mouse models of glioma using single tumor suppressor knockouts or overexpression of single oncogenes mostly failed (Reilly and Jacks, 2001). It was subsequently found that, as confirmed by the TCGA project, the core signaling pathways are crucial for gliomas: genetically engineered mice that activate RTK pathways in the brain, along with simultaneous loss of genes involved in cell-cycle control, develop glioma with high penetrance. Also, like human gliomas, additional loss of the tumor suppressor PTEN causes higher-grade malignancy and reduced survival in mouse glioma models (Kwon et al., 2008). Catalyzed by the profusion of genetic information arising from a number of genome-wide studies that revealed mutations present in human gliomas, as well as advances in molecular biology tools, dozens of genetic mouse glioma models have been generated over the last two decades. Though space limitations prevent us from describing these various models, we have summarized them in Table S1 available online.
Genetic mouse models have been widely used to investigate the cell of origin of malignant glioma (Alcantara Llaguno et al., 2009; Liu et al., 2011; Uhrbom et al., 2005). Recently, Suzanne Baker's group reported the first comprehensive genomic study of a mouse model of high-grade astrocytoma (HGA) generated by manipulating tumor suppressors commonly mutated in human HGAs: Pten, P53, and Rb (Chow et al., 2011). Their studies revealed an astonishing similarity in gene copy number and alteration between mouse HGAs and human GBMs. The mouse tumors also showed similar molecular subtypes as those found in human malignant gliomas (Chow et al., 2011; Verhaak et al., 2010). This study demonstrated the physiological relevance and value of mouse glioma models for future preclinical studies.
As genome-wide sequencing efforts continue in humans, ever more faithful mouse glioma models that better recapitulate the complex genomic landscape of human glioma will be generated. These models will provide increasingly powerful tools for the validation of hypotheses engendered by human genomic data, such as confirming the driver mutations that are causal to oncogenesis, as well as for the preclinical testing of personalized therapy.
Glioma Stem Cells
Another rapidly progressing yet controversial area in glioma research is that of cancer stem cells (CSCs). CSCs, as defined by the American Association for Cancer Research (AACR) workshop on CSCs, are a subpopulation of cells in the tumor that have self-renewal capacity and can give rise to heterogeneous cancer cells that comprise the tumor (Clarke et al., 2006). Glioma stem cells (GSCs) were one of the first such type of cells isolated from solid tumors (Singh et al., 2004). In the original report, as few as 100 GSCs could give rise to tumors that recapitulated the parental tumors when implanted in immunodeficient mice, whereas as many as 1,000,000 non-GSCs could not (Singh et al., 2004).
Unlike traditional glioma cell culture with serum, GSCs were cultured in vitro in monolayer or as spheroids using serum-free medium containing EGF and FGF (Lee et al., 2006; Singh et al., 2004). Interestingly, GSCs isolated from human tumors and cultured in vitro showed remarkable similarities to normal neural stem cells (NSCs), expressing neural stem/progenitor markers such as Nestin, Sox2, and Olig2 and, upon induction, could be differentiated to cells expressing neuronal or glial markers. Using cells derived from the same primary GBM tissues, Lee et al. performed an extensive comparison of traditional serum-containing culture versus serum-free GSC culture (Lee et al., 2006). They reported that the genetic content of parental tumors can be stably preserved under stem cell culture conditions, whereas cells maintained in serum-containing medium underwent dramatic genetic and epigenetic changes over time. More importantly, transplanting GSCs into immunodeficient mice yielded tumors that shared similar histology and global gene expression patterns with their parental tumors. By contrast, in serum-containing medium, early passage glioma cells were completely incapable of tumor formation after transplantation, whereas late-passage glioma cells gave rise to morphologically distinct tumors containing a different molecular signature than the original tumors (Lee et al., 2006). The GSC culture thus provided a more reliable and physiologically relevant model to study disease mechanism.
The discovery of GSCs provided a possible explanation for glioma recurrence following treatment. It was reported that CD133+ glioma stem cells in vitro were more resistant to radiation compared to the CD133– population due to activation of DNA repair pathways by CHK1/CHK2 (Bao et al., 2006a). In addition, GSCs were found to overexpress certain ATP-binding cassette transporters (ABCTs) such as ABCG2 and to export the chemotherapy agent temozolomide (TMZ), a mechanism that was linked to the PTEN/PI3K/AKT pathway (Bleau et al., 2009). The stem cell properties of GSCs also provide new strategies for therapeutic interventions. Numerous studies have shown that pathways such as Sonic hedgehog, Notch, and Wnt, as well as key “stemness” factors such as Olig2, Bmi1, and Nanog, play important roles in GSC maintenance (Bruggeman et al., 2007; Ligon et al., 2007; Po et al., 2010; Stiles and Rowitch, 2008; Zbinden et al., 2010; Zheng et al., 2010). Chemical compounds or RNAi constructs that block components of those pathways have been shown to slow down GSC growth in vitro and attenuate tumor formation in transplantation assays. On the other hand, factors that promote differentiation can be another viable approach to eliminate CSCs. One such example is BMP4, which induces astrocytic differentiation of GSCs and dramatically reduces the tumorigenic potential of CD133+ human GSCs (Piccirillo et al., 2006). Other strategies include targeting stem cell-specific markers, stem cell niches, and quiescent stem cells; these strategies have been summarized in an excellent review article by Zhou et al. (Zhou et al., 2009).
Controversies in the Glioma Stem Cell Field
GSC Markers
The field of somatic stem cells and CSCs has been led by the hematopoietic field, in which relatively large subpopulations of cells that grow in suspension can be discerned and isolated using panels of extracellular epitopes. This permits effective enrichment and study of specific subpopulations of stem cells, progenitor cells, or leukemia subtypes (Bonnet and Dick, 1997). Unfortunately, the solid tumor CSC field has not benefited as well from such epitope-reliant classifications.
Initially, GSCs were cultured as spheroids in EGF- and FGF-containing medium, and it was reported that CD133/prominin-1-positive cells were enriched for CSCs (Singh et al., 2004). However, subsequent studies suggested that CD133 was not a reliable CSC marker: two different groups reported that CD133-negative tumor cells isolated from GBMs can be stably passaged under the same stem cell conditions. Interestingly, similar to the CD133+ cells, these cells also showed “stem cell” properties such as self-renewal and differentiation in vitro and formed transplantable tumors in a xenograft model (Beier et al., 2007; Joo et al., 2008). One study reported that, unlike the CD133+ cells, which can form floating spheroids in culture, the CD133– cells tend to grow as adherent spheres. Recently, a small population of CD133– cells that can give rise to CD133+ cells was reported, suggesting a possible stem cell hierarchy in the spheroid culture system that may or may not have in vivo relevance (Chen et al., 2010). More generally applicable stem cell markers have also been suggested, such as CD44, CD15, and integrin α 6, although they have not been extensively validated by the community (Anido et al., 2010; Lathia et al., 2010; Son et al., 2009).
GSC Niches
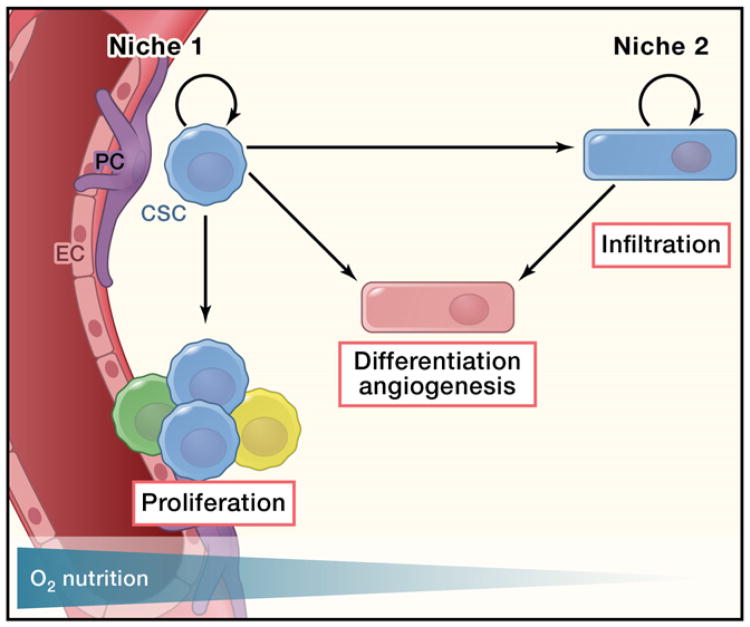
It is known that normal NSCs preferentially reside in specific anatomic regions of the brain: the subventricular zone (SVZ) and subgranular layer (SGZ) (Zhao et al., 2008). In those regions, specific cells, including endothelial and ependymal cells, form a niche that nourishes the NSCs and supports their self-renewal (Shen et al., 2004). This “seed-and-soil” relationship has been adapted in CSC research, as CSCs also require a specific microenvironment to maintain their “stem cell” properties. Like normal NSCs, Nestin+ and CD133+ GSCs have been reported to reside in a vascular niche (Calabrese et al., 2007). Coculturing CD133+ GSCs with endothelial cells enhanced their proliferation in vitro. Coimplantation of both CD133+ and endothelial cells promoted tumor growth in a xenograft model, whereas angiogenesis antagonists eradicated GSCs and significantly slowed tumor growth (Calabrese et al., 2007). However, the in vitro observation from coculture experiments could potentially be compromised by the strong mitogenic effect of growth factors in the cell culture medium, and the putative “stem cell” markers Nestin and CD133 are also expressed by endothelial cells, making the immunohistochemistry results unreliable (Kelly et al., 2007; Klein et al., 2003). Furthermore, a “chicken or egg first” paradox exists in the vascular niche hypothesis, as GSCs themselves can elicit angiogenic effects by secreting factors such as SDF-1 and VEGFA (Bao et al., 2006b; Folkins et al., 2009). Under certain conditions, GSCs can even directly transdifferentiate into the endothelial lineage, making it difficult to determine whether the vascular niche is required for glioma stem cell maintenance (Ricci-Vitiani et al., 2010; Wang et al., 2010). Finally, the vascular niche hypothesis fails to explain several clinical and experimental observations. Malignant glioma cells are notoriously infiltrative and invariably recur after surgical removal of the tumor, suggesting that the peripheral regions of the tumor contain residual tumor cells. Histological observations from both human GBM samples and mouse tumor models suggest that glioma cells typically migrate along white matter (WM) tracts and not their proposed vascular niches (Louis et al., 2007). It is known that hypoxic conditions facilitate self-renewal of both NSCs and GSCs in culture (Clarke and van der Kooy, 2009; Li et al., 2009c). HIF-2α was reported to regulate tumorigenic potential of GSCs, and its expression correlates with poor clinical outcome of glioma patients (Li et al., 2009c). A high percentage of CD133-positive cells was also observed in the pseudopalisade formations that delineate the necrotic center of GBMs, which was thought to be caused by hypoxia (Christensen et al., 2008). In vivo, deletion of HIF-1α resulted in a decline in hippocampal neurogenesis (Mazumdar et al., 2010). It is thus difficult to reconcile the function of hypoxia in neural/glioma stem cells with the vascular niche hypothesis, wherein high levels of oxygen are supplied to neighboring stem cells. Additionally, in several mouse tumor models, inhibition of angiogenesis can shrink the original tumor but also promotes malignant progression by increasing tumor cell invasion and metastasis (Ebos et al., 2009; Pàez-Ribes et al., 2009). Similar findings have been observed in bevacizumab-treated GBM patients in whom the tumor phenotype shifted to a predominantly infiltrative pattern (de Groot et al., 2010; di Tomaso et al., 2011). As a result, bevacizumab treatment leads to prolonged progression-free survival, but not overall survival time (T.F. Cloughesy et al., 2008, ASCO, abstract). Therefore, there could be more than one defined niche for GSCs (Figure 3). A hypoxic environment could be one such niche. Hypoxia promotes GSC (most likely quiescent because of a lack of nutrient supply) infiltration. The infiltrating GSCs, under certain conditions, can home to vascular niches or recruit endothelial cells by secretion of chemokines such as SDF-1. Similar to what has been observed for normal NSCs (Kokovay et al., 2010), the newly established nutritional source then activates the quiescent cancer cells to proliferate and colonize new tumors.
Figure 3. The Vascular Niche and Hypoxic Niche of Glioma Stem Cells.
Vascular niches have been found to be important for glioma growth, probably due to the secreted factors from the endothelial cells within the niche, as well as the nutrient supply from the blood vessels. On the other hand, constantly infiltrating glioma stem cells (GSCs) probably maintain their stemness through activation of hypoxia-related pathways. At the same time, GSCs can recruit endothelial cells by secreting angiogenic factors or by directly differentiating into cells of the endothelial lineage, which in turn support glioma growth. EC and PC are endothelial cell and pericyte cell, respectively.
CSC Assays
CSCs were defined as cells that maintain the tumor in vivo (Clarke et al., 2006). Paradoxically, the field relies entirely on the assessment and study of CSCs by prospective isolation of a subpopulation of cells from tumors and ex vivo culture or tumorigenic assays using immunodeficient rodents (Clarke et al., 2006). Thus, the behavior of the transplanted cancer cells outside of their original environment is studied, and the readout is inferred to reflect how the cells would behave in their original tumor environment. Although the tumorigenic assay of serially transplanting cells into immunodeficient hosts is considered a validation of CSCs, the ability to form a tumor in an immunodeficient host does not necessarily correlate with the implicit definition of a CSC, which is concerned with its presumed role in tumor development and maintenance in vivo (Jordan, 2009; Quintana et al., 2008). In fact, primary glioma cells, as well as traditionally established glioma cell lines, in serum-containing “non-stem cell” culture condition, can form tumors when transplanted. Whereas primary gliomas can be classified into PN, MES, and proliferative subtypes (Phillips et al., 2006), xenograft tumors from glioma cell lines all express high levels of proliferative markers (Hodgson et al., 2009). Recently, it has also been reported that the variability of the transplantation assay could greatly impact the estimation of CSC frequency (Boiko et al., 2010; Quintana et al., 2008). New technologies or model systems are needed to permit direct evaluation of putative GSCs in native tumor development and maintenance.
Finally, CSC nomenclature was adopted from normal tissue stem cells to describe the abilities required for maintaining tumor growth: self-renewal and giving rise to other tumor cells. The CSC concept and properties are frequently confused with the concept and properties of normal tissue stem cells (Jordan, 2009). The hierarchy of normal tissue stem cells is well defined and conserved. In contrast, tumors show a huge degree of diversity. For example, CSCs in mouse models of lung cancer driven by different mutations express distinct surface antigens (Curtis et al., 2010).
Possible Causes Underlying These Controversies
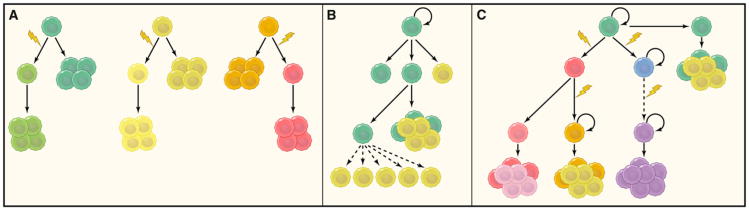
The inconsistent findings observed in GSC research could be due to the fact that gliomas can be classified into several subtypes according to either pathology or molecular genetics. In fact, it was reported that GSC spheroids could not be derived from secondary GBMs (Beier et al., 2007). CD133- GBMs usually show different histology, and the GSCs isolated from these tumors harbor distinct molecular signatures compared to CD133+ tumors (Beier et al., 2007; Joo et al., 2008). Additionally, although spheroid cultures can be established from various brain tumor types in addition to glioma, these cultures can display distinct properties. For example, addition of BMP4 to GSCs isolated from astrocytic tumors and GBM can efficiently differentiate these cells, whereas it has only a minor effect on spheroids isolated from oligodendrogliomas (Persson et al., 2010; Piccirillo et al., 2006). Likewise, even in the same tumor, CSCs are not likely to be a static population: multiple clones of CSCs with diverse genetic alterations could exist, and those stem cells would be under selective pressure at the same time (Figure 4).
Figure 4. Cancer Maintenance Models.
(A) Clonal evolution model. In this model, tumor cells are equivalent, and a majority of the tumor cells have the ability to sustain tumor growth.
(B) Traditional view of the cancer stem cell model. In this model, a stable hierarchy exists in the cells of the tumor, whereby only cancer stem cells have the ability to self-renew and contribute to long-term maintenance of tumor growth.
(C) Evolutional view of cancer stem cell model. This model posits that the hierarchical structure of cancer stem cells is constantly evolving due to natural selection and genomic instability. New cancer stem cell clones with different genetic alterations emerge over time. Certain genetic events will ultimately confer most tumor cells with self-renewal capacity without the reliance of niches or stemness factors (dashed arrows).
It has been suggested that the tumor is maintained by a cellular hierarchy that is similar to Darwin's evolutionary tree structure rather than a linear hierarchical structure (Greaves, 2010). Consistent with this idea, Piccirillo et al. isolated two “stem-like” cell populations from the periphery and the core of the same GBM samples that display distinct tumorigenic potential and cytogenetic profiles (Piccirillo et al., 2009). The heterogeneity of tumor phenotypes is thus likely to be determined by both the clonal diversity of CSCs and their differentiation capacity. Another theory, yet to be demonstrated in vivo, posits that the progeny of CSCs are plastic and can revert back to a stem cell-like state. In melanoma, it was reported that a slow-cycling JARID1B-positive population that can give rise to JARID1B-negative cells is essential for tumor growth and for preventing growth exhaustion (Roesch et al., 2010). However, in contrast to the typical unidirectional hierarchy of cancer stem cells, the JARID1B-expressing population is not predetermined and can be dynamically changed. Using cell line models, Weinberg et al. proposed another hypothesis: that a process similar to epithelial to mesenchymal transition (EMT) could lead to conversion of the non-CSCs and CSC compartments (Mani et al., 2008). EMT is a developmentally regulated process and is believed to mediate breast cancer metastasis; breast epithelial cells that have undergone EMT showed CSC characteristics (Mani et al., 2008). It is known that sphere-forming cells in wild-type SVZ neural stem cell culture are mostly derived from the progenitor cells that respond to EGF and FGF stimulation (Doetsch et al., 2002). Whether similar processes are also involved in glioma development in vivo is still unclear.
Cell of Origin of Glioma
We have discussed the notion that differences in genetic alterations could be responsible for generating glioma subtype diversity. Another potential contributing factor to heterogeneity could be the tumor cell of origin. Cells of origin are the normal cells in which tumorigenic mutations first occur and accumulate to form a full-blown malignancy. CSCs, on the other hand, are defined as the cells that maintain an already formed tumor. Unfortunately, the two concepts are frequently confused, especially when CSCs are also referred to as “tumor-initiating cells.” As Jane E. Visvader pointed out, the term “tumor-initiating cells” is more in line with the “cells of origin,” whereas CSCs would more accurately be referred to as “tumor-propagating cells” (Visvader, 2011).
Prior to the discovery of adult NSCs, astrocytes were thought to be the origin of gliomas, as they were the only known replication-competent population (Figure 5). The malignant transformation process in this scenario requires a “dedifferentiation” process by which differentiated cells regain immature glial and progenitor properties. The feasibility of this dedifferentiation process is supported by recent findings demonstrating that a certain transcription factor cocktail can reprogram terminally differentiated cells back to pluripotent embryonic stem cells (Takahashi and Yamanaka, 2006). Indeed, neonatal cortical astrocytes in culture can be reverted back to a neural stem/progenitor-like status by deleting the tumor suppressors INK4a/ARF or prolonging treatment with growth factors (Bachoo et al., 2002). Several studies reported successful generation of gliomas by transforming early cortical astrocytes in vitro and in vivo (Bachoo et al., 2002; Uhrbom et al., 2005). However, evidence supporting mature astrocytes as possible cells of origin of gliomas is still lacking. In vivo efforts using genetically engineered mice or viral delivery were limited by the lack of a good mature astrocyte marker. It is now well known that the widely used “astrocyte” marker GFAP is also expressed by adult NSCs (Figure 5) (Doetsch et al., 1999). In vitro, culturing mature astrocytes is extremely difficult, and astrocyte cultures from neonatal mouse cortex were reported to contain immature progenitor cells (Laywell et al., 2000). In addition, transformation of neonatal astrocytes relies on deletion of INK4a/ARF in many studies. Given their roles in reprogramming and aging (Li et al., 2009b; Utikal et al., 2009), whether the germline deletion of INK4a/ARF itself affects glial maturation is still unclear and thus further compromises the interpretation of these studies.
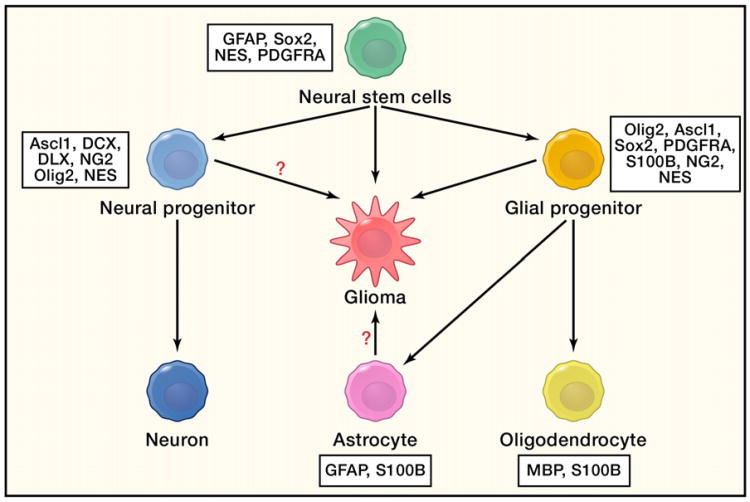
Figure 5. Cell of Origin of Gliomas.
Mutations in adult neural stem cells are sufficient to drive malignant glioma formation in vivo. Some evidence for progenitors or mature astrocytes as the cells of origin has been demonstrated in vitro and in vivo. However, strict proof has not yet been shown due to the lack of specific in vivo lineage markers.
The rediscovery of self-renewing NSCs in postnatal mammalian brains (Zhao et al., 2008) provided an attractive alternative candidate for the glioma cell of origin (Figure 5). Self-renewal capacity gives the NSCs a natural “advantage” in accumulating oncogenic mutations. The oncogenic process can then be viewed as NSCs losing control over their self-renewal and differentiation properties. Several lines of evidence suggest that NSCs are more susceptible to malignant transformation. (1) Abnormalities first occur in the NSC niches of pretumorigenic mice in spontaneous mouse models of glioma (Kwon et al., 2008; Zhu et al., 2005). (2) Temporally deleting the tumor suppressors P53, NF1, and Pten specifically in postnatal mouse neural stem/progenitor cells using a tamoxifen-inducible Nestin-Cre resulted in glioma formation with 100% penetrance, whereas ablation of these genes in nonneurogenic adult brain regions using Cre adenovirus did not produce tumors (Alcantara Llaguno et al., 2009). Similarly, ablation of P53, Pten, and/or Rb in SVZ stem cells, but not in peripheral astrocytes, yielded gliomas (Jacques et al., 2010). It should be noted that these experiments were unable to distinguish between the more quiescent, long-term self-renewing NSCs and the more rapidly dividing progenitor cells, which have limited self-renewal potential. Recently, Liu et al. used mosaic analysis with double markers (MADM)to elegantly demonstrate that the early expanding tumor cell populations in the Nf1;P53-based mouse oligoastrocytoma model are cells that express OPC markers (Liu et al., 2011). In addition, gliomas can be formed by deletion of NF1 and P53 in NG2+ cells, the majority of which are OPCs (Liu et al., 2011). Nevertheless, similar caveats also exist in those studies. Human glioma cells are known to migrate through the WM tracts, and the oligodendrocyte lineage marker PDGFRA is expressed by both NSCs and OPCs (Figure 5) (Jackson et al., 2006). In vivo, NG2 is expressed by multiple cell types, including OPCs, pericytes, and microglial cells (Richardson et al., 2011). A population of multipotent NG2+ cells was also identified in the mouse brain (Aguirre et al., 2004; Rivers et al., 2008; Zhu et al., 2008); it is still unknown whether those cells have long-term self-renewal capacity. Interestingly, lineage tracing of NG2+ cells revealed that they can give rise to both oligodendrocytes and gray matter astrocytes (Zhu et al., 2008). Similarly, a bipotent NG2-positive oligodendrocyte type 2 astrocyte progenitor cell population can be isolated and differentiated to either OPCs or type 2 astrocytes, depending on the medium in vitro. However, the in vivo counterpart of such type 2 astrocytes is still a mystery (Richardson et al., 2011). Finally, SVZ NSCs can differentiate into oligodendrocytes and migrate into the WM (Menn et al., 2006). This process can be greatly enhanced by EGF infusion (Gonzalez-Perez and Alvarez-Buylla, 2011), and amplification of EGFR was frequently identified in both astrocytomas and oligodendro-gliomas (Persson et al., 2010).
Mouse models are important tools for investigating and validating the cell of origin and the natural history of cancers, and it is critical that genetic alterations and physiological setting of tumor development closely resemble that seen in human patients. In addition, cell-of-origin studies address tumor development and progression, which usually takes years if not decades in human patients. An ideal model system for studying the cell of origin should not only address whether the cells are capable of transformation following a set of simultaneous oncogenic events, but also by accumulation of mutations over time. Knowledge of cell-specific markers combined with technologies that achieve more precise temporal and spatial somatic gene manipulation would greatly facilitate future studies. Histopathology and higher-resolution molecular genetic analyses (e.g., single-cell analysis) of patient samples at different stages will also provide valuable information. For example, Lai et al. carefully examined human GBMs containing the IDH1 R132 mutation and observed a number of unique features that distinguished them from other GBMs, including predominantly frontal lobe involvement (Lai et al., 2011). Their conclusion from these phenotypic differences was that the IDH R132 GBMs most likely arise from unique cell types of origin. Microarray studies of human GBM samples revealed that different molecular subtypes share similarity with profiles of different neural lineages, suggesting potentially different origins (Verhaak et al., 2010). However, studies in other solid tumors, including breast cancer, pancreatic tumors, and basal cell carcinomas, reported that histopathology and molecular markers of malignant tumors can be misleading. Breast cancers with BRCA1 mutations usually show basal celllike marker expression even though they arise from luminal progenitor cells (Lim et al., 2009). Also, early abnormalities were observed in the acinar cells from Kras-driven pancreatic duct carcinomas (De La O et al., 2008). It has yet to be determined whether a similar phenomenon occurs in the origin of gliomas.
Conclusions
Although many questions and controversies remain, our understanding of malignant glioma has increased dramatically in recent years. For the first time, we have a clear picture of the human GBM genomic landscape. The continued incorporation and validation of new data using ever more sophisticated animal models will further advance our knowledge of disease origin, progression, and treatment. At the same time, despite the problems outlined, the CSC theory not only points to new cancer targeting strategies from aspects of developmental biology, but also provides insights into tumor maintenance, therapy resistance, and recurrence. The accumulation of this knowledge provides great opportunities to improve and even revolutionize current diagnosis and treatment of human malignant glioma, especially GBM, which typically causes mortality within 1 year.
Supplementary Material
Footnotes
Supplemental Information: Supplemental Information includes one table and can be found with this article online at doi:10.1016/j.cell.2012.03.009.
References
- Aguirre AA, Chittajallu R, Belachew S, Gallo V. NG2-expressing cells in the subventricular zone are type C-like cells and contribute to inter neuron generation in the postnatal hippocampus. J Cell Biol. 2004;165:575–589. doi: 10.1083/jcb.200311141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, Alvarez-Buylla A, Parada LF. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anido J, Sáez-Borderías A, González-Juncà A, Rodón L, Folch G, Carmona MA, Prieto-Sánchez RM, Barba I, Martínez-Sáez E, Prudkin L, et al. TGF-β Receptor Inhibitors Target the CD44(high)/Id1(high) Glioma-Initiating Cell Population in Human Glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- Attolini CS, Cheng YK, Beroukhim R, Getz G, Abdel-Wahab O, Levine RL, Mellinghoff IK, Michor F. A mathematical framework to determine the temporal sequence of somatic genetic events in cancer. Proc Natl Acad Sci USA. 2010;107:17604–17609. doi: 10.1073/pnas.1009117107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radio-resistance by preferential activation of the DNA damage response. Nature. 2006a;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD, Rich JN. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006b;66:7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U, Beier CP. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- Bleau AM, Hambardzumyan D, Ozawa T, Fomchenko EI, Huse JT, Brennan CW, Holland EC. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell. 2009;4:226–235. doi: 10.1016/j.stem.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiko AD, Razorenova OV, van de Rijn M, Swetter SM, Johnson DL, Ly DP, Butler PD, Yang GP, Joshua B, Kaplan MJ, et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 2010;466:133–137. doi: 10.1038/nature09161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, Yu IL, Carro MS, Dai F, Tagge MJ, et al. NFKBIA deletion in glioblastomas. N Engl J Med. 2011;364:627–637. doi: 10.1056/NEJMoa1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman SW, Hulsman D, Tanger E, Buckle T, Blom M, Zevenhoven J, van Tellingen O, van Lohuizen M. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell. 2007;12:328–341. doi: 10.1016/j.ccr.2007.08.032. [DOI] [PubMed] [Google Scholar]
- Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CBTRUS (Central Brain Tumor Registry of the United States) CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in Eighteen States in 2002-2006 2009 [Google Scholar]
- Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, Greve JM, Soriano RH, Gilmour LL, Rivers CS, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Chow LM, Endersby R, Zhu X, Rankin S, Qu C, Zhang J, Broniscer A, Ellison DW, Baker SJ. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell. 2011;19:305–316. doi: 10.1016/j.ccr.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Schrøder HD, Kristensen BW. CD133 identifies perivascular niches in grade II-IV astrocytomas. J Neurooncol. 2008;90:157–170. doi: 10.1007/s11060-008-9648-8. [DOI] [PubMed] [Google Scholar]
- Clarke L, van der Kooy D. Low oxygen enhances primitive and definitive neural stem cell colony formation by inhibiting distinct cell death pathways. Stem Cells. 2009;27:1879–1886. doi: 10.1002/stem.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells— perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- Curtis SJ, Sinkevicius KW, Li D, Lau AN, Roach RR, Zamponi R, Woolfenden AE, Kirsch DG, Wong KK, Kim CF. Primary tumor genotype is an important determinant in identification of lung cancer propagating cells. Cell Stem Cell. 2010;7:127–133. doi: 10.1016/j.stem.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465:966. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot JF, Fuller G, Kumar AJ, Piao Y, Eterovic K, Ji Y, Conrad CA. Tumor invasion after treatment ofglioblastoma with bevacizumab: radiographic and pathologic correlation in humans and mice. Neuro-oncol. 2010;12:233–242. doi: 10.1093/neuonc/nop027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La O JP, Emerson LL, Goodman JL, Froebe SC, Illum BE, Curtis AB, Murtaugh LC. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci USA. 2008;105:18907–18912. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Tomaso E, Snuderl M, Kamoun WS, Duda DG, Auluck PK, Fazlollahi L, Andronesi OC, Frosch MP, Wen PY, Plotkin SR, et al. Glioblastoma recurrence after cediranib therapy in patients: lack of “rebound” revascularization as mode of escape. Cancer Res. 2011;71:19–28. doi: 10.1158/0008-5472.CAN-10-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron. 2002;36:1021–1034. doi: 10.1016/s0896-6273(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Ebos JM, Lee CR, Cruz-Munoz W, Bjarnason GA, Christensen JG, Kerbel RS. Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell. 2009;15:232–239. doi: 10.1016/j.ccr.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkins C, Shaked Y, Man S, Tang T, Lee CR, Zhu Z, Hoffman RM, Kerbel RS. Glioma tumor stem-like cells promote tumor angiogenesis and vasculogenesis via vascular endothelial growth factor and stromal-derived factor 1. Cancer Res. 2009;69:7243–7251. doi: 10.1158/0008-5472.CAN-09-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez O, Alvarez-Buylla A. Oligodendrogenesis in the subventricular zone and the role of epidermal growth factor. Brain Res Rev. 2011;67:147–156. doi: 10.1016/j.brainresrev.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravendeel LAM, Kouwenhoven MCM, Gevaert O, de Rooi JJ, Stubbs AP, Duijm JE, Daemen A, Bleeker FE, Bralten LBC, Kloosterhof NK, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69:9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- Greaves M. Cancer stem cells: back to Darwin? Semin Cancer Biol. 2010;20:65–70. doi: 10.1016/j.semcancer.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Green A, Beer P. Somatic mutations of IDH1 and IDH2 in the leukemic transformation of myeloproliferative neoplasms. N Engl J Med. 2010;362:369–370. doi: 10.1056/NEJMc0910063. [DOI] [PubMed] [Google Scholar]
- Gutmann DH, Rasmussen SA, Wolkenstein P, MacCollin MM, Guha A, Inskip PD, North KN, Poyhonen M, Birch PH, Friedman JM. Gliomas presenting after age 10 in individuals with neurofibromatosis type 1 (NF1) Neurology. 2002;59:759–761. doi: 10.1212/wnl.59.5.759. [DOI] [PubMed] [Google Scholar]
- Hodgson JG, Yeh RF, Ray A, Wang NJ, Smirnov I, Yu M, Hariono S, Silber J, Feiler HS, Gray JW, et al. Comparative analyses of gene copy number and mRNA expression in glioblastoma multiforme tumors and xenografts. Neuro-oncol. 2009;11:477–487. doi: 10.1215/15228517-2008-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JM. Other Malignancies. In: Friedman JM, Gutmann DH, MacCollin M, Riccardi VM, editors. Neurofibromatosis: Phenotype, Natural History, and Pathogenesis. Baltimore, MD: Johns Hopkins Press; 1999. p. 19. [Google Scholar]
- Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, Roy M, Quinones-Hinojosa A, VandenBerg S, Alvarez-Buylla A. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51:187–199. doi: 10.1016/j.neuron.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Jacques TS, Swales A, Brzozowski MJ, Henriquez NV, Linehan JM, Mirzadeh Z, O' Malley C, Naumann H, Alvarez-Buylla A, Brandner S. Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes. EMBO J. 2010;29:222–235. doi: 10.1038/emboj.2009.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurol. 2010;9:717–726. doi: 10.1016/S1474-4422(10)70105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo KM, Kim SY, Jin X, Song SY, Kong DS, Lee JI, Jeon JW, Kim MH, Kang BG, Jung Y, et al. Clinical and biological implications of CD133-positive and CD133-negative cells in glioblastomas. Lab Invest. 2008;88:808–815. doi: 10.1038/labinvest.2008.57. [DOI] [PubMed] [Google Scholar]
- Jordan CT. Cancer stem cells: controversial or just misunderstood? Cell Stem Cell. 2009;4:203–205. doi: 10.1016/j.stem.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- Kelly PN, Dakic A, Adams JM, Nutt SL, Strasser A. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317:337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- Klein T, Ling Z, Heimberg H, Madsen OD, Heller RS, Serup P. Nestin is expressed in vascular endothelial cells in the adult human pancreas. J Histochem Cytochem. 2003;51:697–706. doi: 10.1177/002215540305100601. [DOI] [PubMed] [Google Scholar]
- Kokovay E, Goderie S, Wang Y, Lotz S, Lin G, Sun Y, Roysam B, Shen Q, Temple S. Adult SVZ lineage cells home to and leave the vascular niche via differential responses to SDF1/CXCR4 signaling. Cell Stem Cell. 2010;7:163–173. doi: 10.1016/j.stem.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranendijk M, Struys EA, van Schaftingen E, Gibson KM, Kanhai WA, van der Knaap MS, Amiel J, Buist NR, Das AM, de Klerk JB, et al. IDH2 mutations in patients with D-2-hydroxyglutaric aciduria. Science. 2010;330:336. doi: 10.1126/science.1192632. [DOI] [PubMed] [Google Scholar]
- Kwon CH, Zhao D, Chen J, Alcantara S, Li Y, Burns DK, Mason RP, Lee EY, Wu H, Parada LF. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res. 2008;68:3286–3294. doi: 10.1158/0008-5472.CAN-07-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A, Kharbanda S, Pope WB, Tran A, Solis OE, Peale F, Forrest WF, Pujara K, Carrillo JA, Pandita A, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol. 2011;29:4482–4490. doi: 10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Gallagher J, Heddleston JM, Wang J, Eyler CE, Macswords J, Wu Q, Vasanji A, McLendon RE, Hjelmeland AB, Rich JN. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latini A, Scussiato K, Rosa RB, Llesuy S, Belló-Klein A, Dutra-Filho CS, Wajner M. D-2-hydroxyglutaric acid induces oxidative stress in cerebral cortex of young rats. Eur J Neurosci. 2003;17:2017–2022. doi: 10.1046/j.1460-9568.2003.02639.x. [DOI] [PubMed] [Google Scholar]
- Laywell ED, Rakic P, Kukekov VG, Holland EC, Steindler DA. Identification of a multipotent astrocytic stem cell in the immature and adult mouse brain. Proc Natl Acad Sci USA. 2000;97:13883–13888. doi: 10.1073/pnas.250471697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Li A, Walling J, Ahn S, Kotliarov Y, Su Q, Quezado M, Oberholtzer JC, Park J, Zenklusen JC, Fine HA. Unsupervised analysis of transcriptomic profiles reveals six glioma subtypes. Cancer Res. 2009a;69:2091–2099. doi: 10.1158/0008-5472.CAN-08-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Cañamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009b;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009c;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. kConFab. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, Zong H. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher EA, Brennan C, Wen PY, Durso L, Ligon KL, Richardson A, Khatry D, Feng B, Sinha R, Louis DN, et al. Marked genomic differences characterize primary and secondary glioblastoma subtypes and identify two distinct molecular and clinical secondary glioblastoma entities. Cancer Res. 2006;66:11502–11513. doi: 10.1158/0008-5472.CAN-06-2072. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelialmesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar J, O'Brien WT, Johnson RS, LaManna JC, Chavez JC, Klein PS, Simon MC. O2 regulates stem cells through Wnt/β-catenin signalling. Nat Cell Biol. 2010;12:1007–1013. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- Menn B, Garcia-Verdugo JM, Yaschine C, Gonzalez-Perez O, Rowitch D, Alvarez-Buylla A. Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci. 2006;26:7907–7918. doi: 10.1523/JNEUROSCI.1299-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Cancer Genome Atlas Research Network. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pàez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Viñals F, Inoue M, Bergers G, Hanahan D, Casanovas O. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15:220–231. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson AI, Petritsch C, Swartling FJ, Itsara M, Sim FJ, Auvergne R, Goldenberg DD, Vandenberg SR, Nguyen KN, Yakovenko S, et al. Non-stem cell origin for oligodendroglioma. Cancer Cell. 2010;18:669–682. doi: 10.1016/j.ccr.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, Brem H, Olivi A, Dimeco F, Vescovi AL. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- Piccirillo SGM, Combi R, Cajola L, Patrizi A, Redaelli S, Bentivegna A, Baronchelli S, Maira G, Pollo B, Mangiola A, et al. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene. 2009;28:1807–1811. doi: 10.1038/onc.2009.27. [DOI] [PubMed] [Google Scholar]
- Po A, Ferretti E, Miele E, De Smaele E, Paganelli A, Canettieri G, Coni S, Di Marcotullio L, Biffoni M, Massimi L, et al. Hedgehog controls neural stem cells through p53-independent regulation of Nanog. EMBO J. 2010;29:2646–2658. doi: 10.1038/emboj.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly KM, Jacks T. Genetically engineered mouse models of astrocytoma: GEMs in the rough? Semin Cancer Biol. 2001;11:177–191. doi: 10.1006/scbi.2000.0375. [DOI] [PubMed] [Google Scholar]
- Reitman ZJ, Parsons DW, Yan H. IDH1 and IDH2: not your typical oncogenes. Cancer Cell. 2010;17:215–216. doi: 10.1016/j.ccr.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Pallini R, Biffoni M, Todaro M, Invernici G, Cenci T, Maira G, Parati EA, Stassi G, Larocca LM, De Maria R. Tumour vascularization via endothelial differentiation of glioblastoma stemlike cells. Nature. 2010;468:824–828. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Young KM, Tripathi RB, McKenzie I. NG2-glia as multipotent neural stem cells: fact or fantasy? Neuron. 2011;70:661–673. doi: 10.1016/j.neuron.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, Richardson WD. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat Neurosci. 2008;11:1392–1401. doi: 10.1038/nn.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Goderie SK, Jin L, Karanth N, Sun Y, Abramova N, Vincent P, Pumiglia K, Temple S. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 2004;304:1338–1340. doi: 10.1126/science.1095505. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkel-man RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles CD, Rowitch DH. Glioma stem cells: a midterm exam. Neuron. 2008;58:832–846. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- TCGA (The Cancer Genome Atlas Research Network) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso CL, Freije WA, Day A, Chen Z, Merriman B, Perlina A, Lee Y, Dia EQ, Yoshimoto K, Mischel PS, et al. Distinct transcription profiles of primary and secondary glioblastoma subgroups. Cancer Res. 2006;66:159–167. doi: 10.1158/0008-5472.CAN-05-0077. [DOI] [PubMed] [Google Scholar]
- Uhrbom L, Kastemar M, Johansson FK, Westermark B, Holland EC. Cell type-specific tumor suppression by Ink4a and Arf in Krasinduced mouse gliomagenesis. Cancer Res. 2005;65:2065–2069. doi: 10.1158/0008-5472.CAN-04-3588. [DOI] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- Vitucci M, Hayes DN, Miller CR. Gene expression profiling of gliomas: merging genomic and histopathological classification for personalised therapy. Br J Cancer. 2011;104:545–553. doi: 10.1038/sj.bjc.6606031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Chadalavada K, Wilshire J, Kowalik U, Hovinga KE, Geber A, Fligelman B, Leversha M, Brennan C, Tabar V. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468:829–833. doi: 10.1038/nature09624. [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbinden M, Duquet A, Lorente-Trigos A, Ngwabyt SN, Borges I, Ruiz i Altaba A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 2010;29:2659–2674. doi: 10.1038/emboj.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Ying H, Wiedemeyer R, Yan H, Quayle SN, Ivanova EV, Paik JH, Zhang H, Xiao Y, Perry SR, et al. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell. 2010;17:497–509. doi: 10.1016/j.ccr.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BBS, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anti-cancer drug discovery. Nat Rev Drug Discov. 2009;8:806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- Zhu X, Bergles DE, Nishiyama A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development. 2008;135:145–157. doi: 10.1242/dev.004895. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Guignard F, Zhao D, Liu L, Burns DK, Mason RP, Messing A, Parada LF. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005;8:119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.