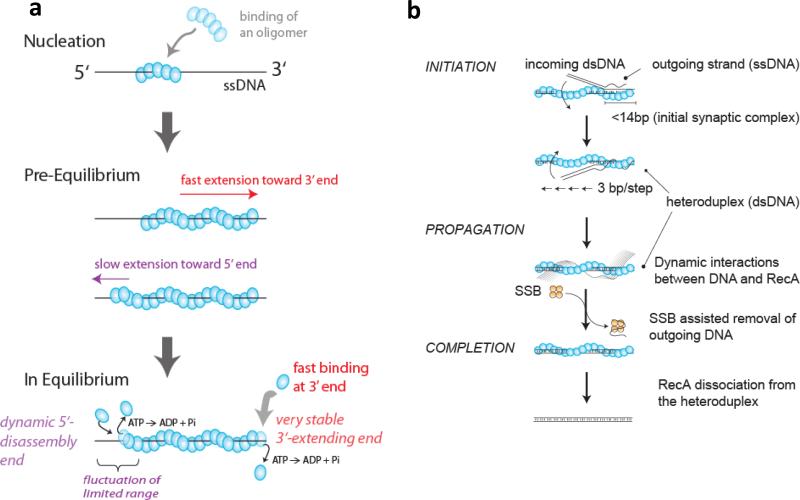
Figure 6. Dynamics of RecA filament formation and strand exchange reactions.
(a) RecA filament formations on ssDNA is nucleated by the binding of a RecA oligomer, likely between 4-5 monomers(37; 59), and is extended rapidly in monomer un units mainly in the 3’ direction with a 10 times slower extension in the 5’ direction(59). In the steady state, RecA monomers can dissociate from both ends as monomers upon ATP hydrolysis(59). (b) RecA-ssDNA filament binds to a homologous dsDNA forming an initial synaptic complex (< 14 bp in length), then propagates rapidly in 3 bp increments while undergoing base pair exchange. The outgoing ssDNA displays dynamic interactions with the resulting RecA-dsDNA filament and its removal is facilitated by SSB. Finally, RecA dissociates from the heteroduplex product, completing the reaction (Adapted from Ref. (105)).