Abstract
Persistent activation of Survivin and its overexpression contribute to the formation, progression and metastasis of several different tumor types. Therefore, Survivin is an ideal target for RNA interference mediated-growth inhibition. Blockade of Survivin using specific short hairpin RNAs (shRNA) can significantly reduce prostate tumor growth. RNA interference does not fully ablate target gene expression, owing to the idiosyncrasies associated with shRNAs and their targets. To enhance the therapeutic efficacy of Survivin-specific shRNA, we employed a combinatorial expression of Survivin-specific shRNA and gene associated with retinoid-interferon-induced mortality-19 (GRIM-19). Then, the GRIM-19 coding sequences and Survivin-specific shRNAs were used to create a dual expression plasmid vector and were carried by an attenuated strain of Salmonella enteric serovar typhimurium (S. typhimurium) to treat prostate cancer in vitro and in vivo. We found that the co-expressed Survivin-specific shRNA and GRIM-19 synergistically and more effectively inhibited prostate tumor proliferation and survival, when compared with treatment with either single agent alone in vitro and in vivo. This study has provided a novel cancer gene therapeutic approach for prostate cancer.
Keywords: GRIM-19, prostate cancer, RNAi, Salmonella enterica serovar typhimurium, Survivin, tumor cell growth
Introduction
Prostate carcinoma is a leading cause of cancer-related deaths of males in Western countries.1 Although we do not have a complete understanding of the molecular mechanisms involved in prostate cancer (PCa) initiation or progression, significant progress has been made in understanding certain gene expression changes that occur in these tumors.2 Some of these gene products have been used for developing antitumor target therapy. For example, Survivin, a member of the inhibitor of apoptosis protein family, is involved in both control of cell division and inhibition of apoptosis.3 Survivin is expressed in embryonic and fetal tissues but is not detected in most normal adult tissues.4 Survivin is overexpressed in almost all human cancers, which makes Survivin one of the top four ‘transcriptomes' expressed in cancer cells in genome-wide searches.5 Previous data indicated that Survivin is associated with cancer progression, drug and radiation resistance and poor outcomes.6 Because of its roles in reducing apoptosis, and stimulating cell division and proliferation, and its differential expression in cancers compared to normal tissues, Survivin is an attractive target for cancer gene therapy.7 Thus far, different approaches have been taken to target Survivin, such as antisense oligonucleotides,8 small interfering RNAs,9 dominant negative mutants,10 ribozymes11 and triplex DNA-dependent kinase for cancer therapeutics.12 However, none of these studies aiming at suppression of Survivin transcription can provide an ideal therapeutic approach. Because of the multiple functions of Survivin, inhibition of its transcription could be an important approach to suppress cancer growth.13
In recent studies, we found that blockade of Survivin using hairpin RNA (shRNA) expression vectors via direct injection can significantly reduce prostate tumor growth in nude mice (data not shown). However, we did not observe complete tumor growth suppression. This observation is in agreement with the fact that RNA interference does not completely block gene transcription, especially when the target mRNA is expressed at abnormally high levels.14, 15 In order to enhance the antitumor efficiency, we added a second anti-tumor gene associated with retinoid-interferon (IFN)-induced mortality (GRIM-19), by constructing a co-expression plasmid.
GRIM-19 was originally isolated using a genetic screen. It is a growth suppressive gene product in the IFN and retinoic acid-induced cell death pathway.16 Accumulating data has revealed that a GRIM-19 mutation plays a role in the development/progression of thyroid carcinoma plus the low expression of GRIM-19 has been observed in some primary human renal and PCas.17, 18 Accumulating data indicated that most tumors express high levels of signal transducer and activator of transcription 3 (Stat3), which is known to upregulate the expression of Survivin.19, 20, 21 Such upregulation in the absence of GRIM-19 may promote tumor growth and metastasis. We show here, for the first time, that co-expression of Survivin-shRNA and GRIM-19 protein from the same plasmid causes a synergistic suppression of prostate tumor growth in vitro and in vivo.
Salmonella enteric serovar typhimurium (S. typhimurium) is an intracellular bacterium, which can survive in macrophages because of its facultative anaerobic growth ability.22 These features allow S. typhimurium to accumulate and multiply in tumor cells. Previous data indicated that attenuated Salmonella selectively gathered in solid tumors at a level 1000 times that observed in normal tissues. Nguyen et al.23 suggested that attenuated Salmonella is the most promising bacterial carrier for the treatment of malignant tumor and other diseases. Attenuated S. typhimurium as a gene delivery vector provides more advantages when compared to viral vectors, such as good targeting.24, 25, 26 For example, Salmonella can: (i) specifically colonize in hypoxic tumor tissues if administered through the intravenous or intraperitoneal routes; (ii) be used across a wide range of tissues; (iii) express foreign genes in any hypoxic region, unlike a retroviral vector which can only infect dividing cells; (iv) proliferate in vivo and efficiently express exogenous gene products; and by itself exerts anti-tumor effects; (v) be an ideal and safe vehicle for gene transfer since that it is sensitive to antibiotics and small doses of currently available antibiotics can kill it; and (vi) not be rejected by the host upon long-term use. Taken together, these characteristics make S. typhimurium an ideal vector for gene therapy.
So we tempted to achieve the superior antitumor effect in vivo which used attenuated Salmonella as barrier to deliver Survivin shRNA and GRIM-19 protein from the same plasmid backbone in this study.
Materials and methods
Immunohistochemical analyses
Thirty-two total prostate tumor samples and 38 normal or hyperplasic prostate tissues were collected for determination of Survivin and GRIM-19 expressions. Immunostaining was performed using Vectastain Elite ABC avidin-biotin staining kit (Vector Labs, Inc., Burlingame, CA, USA). Antibodies specific for Survivin were obtained from Santa Cruz Biotech, Inc. (Santa Cruz, CA, USA). A mouse monoclonal antibody against GRIM-19 was described earlier.27 The criteria for grading immunohistochemical data for Survivin and GRIM-19 were similar to those described earlier.28
Semiquantitative reverse transcription-PCR (RT-PCR)
mRNA levels were determined using semiquantitative RT-PCR. For RT-PCR analysis, total RNA (5 µg) of each sample was reverse-transcribed into complementary DNA using an RT-PCR kit (final volume 20 μl containing: 2 μl 10× buffer, 2 μl 200 μmol l−1 deoxynucleotide triphosphate, 0.2 μl Taq DNA, 1.5 μl 1.5 mmol l−1 Mg2+) (Promega, Madison, WI, USA). The following primer pairs: Sur-sense: 5′-GAATTCATGGGTGCCCCGACGTTGCC-3′ and Sur-antisense: 5′-AGATCTTTCTTCTTATTGTTGGTTTCC-3′ which yield a 475-bp PCR product; and G-19 sense 5′-GAGTCACGCACTGTCTGGG-3′ and G19-antisense 5′-CGGTCGGTTTCTGCCTGTA-3′, which yield a 485-bp PCR product were employed for determining the Survivin and GRIM-19 levels, respectively. The PCR reactions were performed as follows: initial denaturation was 94 °C for 5 min followed by 25 cycles of amplification (94 °C for 30 s, 59 °C for 45 s, 72 °C for 90 s and a final extension for 10 min at 72 °C). The reaction products were analyzed by 1.5% standard agarose gel electrophophoresis with 5 μg ml−1 ethidium bromide (EB) and visualized by a GIS Gelatum imaging system (Tanon, Shanghai, China).
Western blotting
Tissue and cell lyses, protein quantification and western blotting analyses were carried out as described previously.16 Proteins of interest were detected using Survivin and β-actin antibodies from Santa Cruz Biotech according to the manufacture's protocol. GRIM-19 antibody was applied as before.27 One percent of bovine serum albumin was used as control antibody. The absorbance of each band in western blots was quantified using densitometry, and the results are shown as relative expression of each protein from different samples.
Construction of recombinant plasmids
We constructed a series of expression plasmids that expressed an shRNA specific to Survivin and GRIM-19 protein, either alone or in combination, as described previously.29 A Survivin shRNA, which proved to be the most effective at inhibiting growth in our previous study (the sequence of which GCAGTTTGAAGAATTAACC corresponds to nucleotides 394–412; Genbank accession number NM_001168), was used for this study. A negative control Scramble shRNA sequence (Ambion, Austin, TX, USA), which has no significant homology to mouse or human gene sequences, was used as control to demonstrate the specific effects. The oligonucleotide contained a sense strand of 19 nucleotides followed by a short spacer (TTCAAGAGA), the antisense strand, five Ts (terminators), as well as terminal BamHI and HindIII restriction sites. This oligonucleotide construct was cloned into the BamHI and HindIII sites of plasmid pSilencer to generate a Survivin-specific shRNA expression vector, designated psi-Survivin. The U6 promoter and the sequence of Survivin-shRNA or Scramble-shRNA were then PCR amplified with primers: P1, 5′-GC(AGATCT)TGCTTCGCGATGTACGGGCC-3′ P2, 5′-CG(TCGCGA)GGGCTATGAACTAATGACCC-3′, then cloned into T vector to construct plasmids. The human GRIM-19 open reading frame was PCR amplified using the pCXN2mycA GRIM-19 plasmid27 as a template with primers P3, 5′-AGGTACCATGGCGGCGTCAAAGGTGA-3′ and P4, 5′-ACGAATTCCTACGAATTCCTACGTGTACCACATGAAGCC-3′, then cloned into KpnI and EcoRI sites of plasmid pcDNA3.1(+) to construct a plasmid pGRIM-19, in which the GRIM-19 gene was placed under the control of a cytomegalovirus enhancer. T vector was digested with BglII and NurI enzymes and cloned into BglII and NurI sites of pGRIM-19 to generate pGRIM-19-si-Survivin, which could co-express both shRNA-Survivin and protein GRIM-19.
Bacteria, cell culture and transfection
The attenuated S. typhimurium phoP/phoQ null strain LH430 was kindly provided by Dr EL Hohmann.30 The human PCa cell line DU145 was obtained from ATCC (American Type Culture Collection, Manassas, VA, USA) and grown in RPMI-1640 (Life Technologies, Inc, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum. For transfection, cells were plated into a 100-mm dish (106 cells) and allowed to adhere for 24 h. LipofectAMINE 2000 (Invitrogen, Carlsbad, CA, USA) was used for transfection. Quantification of specific mRNA was performed essentially as described previously.29 Briefly, 72 h after transfection, cells were collected and total RNA was extracted using the Trizol reagent (Invitrogen). For RT-PCR analysis, 5 μg of total RNA were subjected to reverse transcription using an RT-PCR kit (Promega). DNAse- I treated RNA was used for cDNA preparation. For amplifying Survivin and GRIM-19 mRNA, the primers shown above were used. Other primers are described in Supplementary Table 1.
Growth assays and apoptosis assays in vitro
DU145 cells (2×105) were plated into 96-well dishes. Cell proliferation was assessed using a commercially available MTT kit (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (Sigma, St Louis, MO, USA) as described previously.31 To detect apoptosis, DU145 cells were incubated in 500 µl of a binding buffer containing of FITC-conjugated annexin V (Clontech Laboratories Inc., Palo Alto, CA, USA) and propidium iodide (PI) at room temperature for 15 min. The samples were then centrifuged at 1000g for 10 min. The supernatant was removed, and the cell pellets were suspended into 500 µl binding buffer and analyzed by flow cytometry. Early apoptotic cells were positive for PI-negative staining and annexin V-positive staining were enumerated. For a negative control, we used mock-infected DU145 cells. Five replicates were performed for each time point. In some experiments, cells were stained with 20 µl acridine orange (AO) after 48h transfection. Cell morphology was observed and photomicrographs were captured.
Antibodies and western blotting
Cleaved caspase3, B-cell lymphoma xL (BcL-xL), c-Myc, Stat3, CyclinD1 and β-actin antibodies were obtained from Santa Cruz Biotech. For western blot analyses, cells were harvested 72 h after transfection and lysed. For western blot analyses, ∼50 µg of total protein from each sample were separated on a 10% SDS–polyacrylamide gel, then transferred onto a PVDF membrane (Millipore, Bedford, MA, USA), and incubated with the indicated antibodies. Protein bands were visualized by the SuperSignal West Pico Chemiluminescent Substrate kit (Pierce Biotechnology, Rockford, IL, USA). Bands corresponding to the proteins of interest were quantified by volume densitometry using the ImageQuant software (Molecular Dynamics, Sunnyvale, CA, USA). The results are presented in comparison to mock transfected control. All experiments were conducted on three separate occasions in duplicate and the results represent the mean±s.e.
Antitumor activity of recombinant attenuated S. typhimurium carrying different plasmids on established prostate tumors
To determine the effects of co-expressed GRIM-19 and Survivin-shRNA on tumor growth we used a tumor xenograft model in nude mice. Mice were transplanted subcutaneously with 2×106 DU145 cells on the right flank. Once palpable tumors (mean diameter, 5 mm) had developed at the sites of injection, mice (n=5 per group) were injected intraperitoneally with 1×107 colony-forming units (CFU) of attenuated S. typhimurium carrying different plasmids. Six experimental groups (five mice per group) were tested: (i) mock transfection (phosphate buffered saline alone); (ii) attenuated Salmonella only (1×107 CFU); (iii) Scramble shRNA control (1×107 CFU of attenuated S. typhimurium with Scramble shRNA vector); (iv) GRIM-19 (1×107 CFU of attenuated S. typhimurium with pGRIM-19 vector); (v) Survivin shRNA (1×107 CFU of attenuated Salmonella with psi-Survivin vector); and (vi) pGRIM-19-si-Survivin (1×107 CFU of attenuated Salmonella with the pGRIM-19-si-Survivin vector). This process was repeated after a week. Mice were euthanized on 49 days after treatments. The tumors were excised for hematoxylin and eosin (H&E) staining, Ki-67 immunohistochemical staining and terminal deoxynucleotidyl transferase-mediated nick end labeling (TUNEL) assay.
H&E staining and TUNEL assays
Tumors excised from mice were processed for H&E staining and TUNEL assays as described previously.28 To quantify apoptosis, 400 nuclei from random microscopic fields were analyzed by an observer blinded to the treatment groups, at least 10 individual sections were evaluated per slide. The total number of apoptotic cells in each section was calculated and expressed as the percentage of the total cell number.
Immunohistochemistry (IHC) staining
Tumor proliferation was determined by digital image analysis with an Automated Cellular Imaging System (ACIS III; DakoCytomation, San Juan Capistrano, CA, USA). Detection of the Ki-67 was determined by IHC on 5-µm-thick formalin-fixed, paraffin embedded sections as described in our earlier studies.28 After removal, paraffin slides were incubated with a 1∶100 dilution of Ki-67-specific antibody for 30 min (positive control slice was applied by Nanjing Jiancheng Bioengineering Institute (Nanjing, China); 1% bovine serum albumin was substituted for first antibody as negative control.). After rinsing, secondary antibody incubation was performed in the DUAL+/HRP-labeled polymer (K4061; DAKO, Carpenteria, CA, USA) for 15 min. Sections were then incubated with 3,3′-diaminobenzidine (DAB+) (K3467; DAKO) for 5 min, counterstained with modified Schmidt's hematoxylin for 5 min and mounted with a permanent mounting media.
Statistical analysis
Quantitative data were expressed as mean±s.e. χ2 analysis was performed to evaluate the significance of intergroup differences. Paired t-test was used for single comparison between two groups. P<0.05 was deemed statistically significant. Statistical calculations were performed using SigmaStat statistical software package (SPSS10.0; SPSS, Chicago, IL, USA).
Results
Survivin and GRIM-19 expression in normal prostate (NP), benign prostatic hyperplasia (BPH), PCa tissues and DU145 prostatic cells
Since GRIM-19 and Survivin exhibit opposite effects on cell growth and proliferation, we first investigated if an inverse correlation exists between the expressions of both proteins in primary tumors versus NP tissues. We screened 32 primary prostate tumors and 16 NP tissues for the expression of Survivin and GRIM-19 using RT-PCR, immunohistochemical and Western blot analyses. The results indicated a low basal expression of Survivin and generally high levels of GRIM-19 in NP tissues. In contrast, primary prostate tumor tissue showed moderate to high expression of Survivin and very low levels of GRIM-19. Immunostaining patterns revealed that 80% of the tumors were positive for moderate to high levels of Survivin, and an extremely low level of GRIM-19 in the tumors(P<0.001) (Supplementary Figure 1a). There were stark differences in GRIM-19 expression between BPH versus PCa (P<0.001). Thus, it appears that some loss of GRIM-19 expression can be found in the early stages of prostatic cancer development. The overexpression of Survivin in these tumors may be due to an autoregulation of its own synthesis or a result of a loss of control over Stat3 (by GRIM-19), which is known to induce Survivin.32 Similarly, RT-PCR and Western blot results also showed that Survivin was highly upregulated in PCa tissue and prostatic DU145 cells while GRIM-19 was severely depressed (P<0.01) (Supplementary Figure 1b–e). The expression levels of Survivin and GRIM-19 in tumors were significantly different from those found in normal tissues (Supplementary Tables 2 and 3). Together these data revealed an inverse correlation between GRIM-19 and Survivin expression in the prostatic carcinomas, which was significantly different from primary prostate tumors. The DU145 cell line also exhibited a similar pattern of GRIM-19 and Survivin expression compared to the normal prostatic tissues, suggesting its usefulness for further studies.
Co-expression of pGRIM-19-si-Survivin in DU145 cells
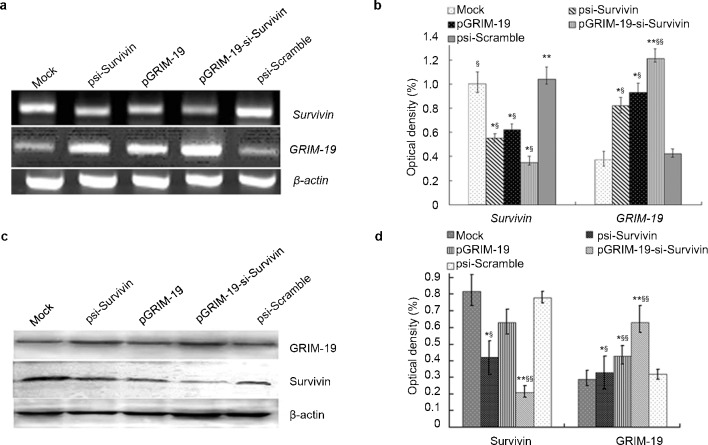
Since plasmid-based Survivin-specific shRNAs alone had an incomplete antitumor effect,14 we investigated if these shRNAs could be complemented with GRIM-19 to achieve maximal growth suppression. Therefore, we constructed a set of expression vectors that could express GRIM-19 and shRNA specific to Survivin individually or in combination. These plasmids were transfected into DU145 cells. Both psi-Scramble and psi-Survivin plasmids carried the green fluorescent protein gene that allowed detection in transfected cells (72.32%±3.89%). We next determined the expression of Survivin and GRIM-19 using RT-PCR and Western blot. RT-PCR results for GRIM-19 and Survivin mRNAs revealed a significant increase in GRIM-19 expression after transfection with expression vectors pGRIM-19 and pGRIM-19-si-Survivin; and a downregulation of Survivin upon transfection with psi-Survivin and pGRIM-19-si-Survivin plasmids (Figure 1a and b). Similar results were obtained with Western blot analyses for GRIM-19 and Survivin proteins (Figure 1c and d).
Figure 1.
Survivin and GRIM-19 mRNA and corresponding protein levels in DU145 cells transfected with recombinant treatment plasmids. (a) RT-PCR analysis of Survivin and GRIM-19 mRNA in DU145 cells transfected with recombinant plasmids. (b) Quantified Survivin and GRIM-19 mRNA levels in DU145 cells transfected with different plasmids. (c) Survivin and GRIM-19 protein level in DU145 cells transfected with different plasmids revealed by western blot. (d) The quantified GRIM-19 and Survivin protein level. Optical density was expressed as mean±s.e. (*P<0.05, **P<0.01 versus mock; §P<0.05, §§P<0.01 versus psi-Scramble). GRIM-19, gene associated with retinoid-interferon-induced mortality-19; RT-PCR, reverse transcription PCR.
Co-expressed Survivin-specific shRNA and GRIM-19 synergistically inhibit growth and promote apoptosis of DU145 cells
To determine whether pGRIM-19-si-Survivin had an inhibitory effect on DU145 cell growth, we employed cell growth assays using MTT (Figure 2a). Although Survivin specific shRNA and GRIM-19 alone significantly suppressed DU145 cell growth, which was better augmented when both of them were co-expressed.
Figure 2.
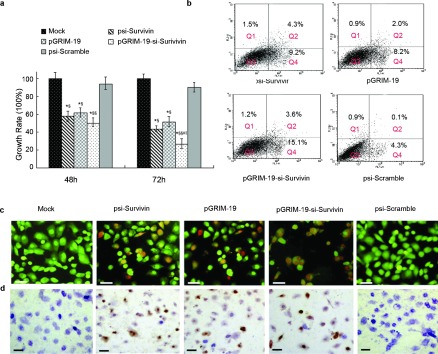
Growth inhibition and apoptosis assays in DU145 cells transfected with different treatment plasmids. (a) Growth inhibitory rate for DU145 cells transfected with psi-Survivin, pGRIM-19, pGRIM-19-si-Survivin and psi-Scramble (*P<0.05, **P<0.01 versus mock; §P<0.05, §§P<0.01 versus psi-Scramble group; #P<0.05 versus psi-Survivin; ‖‖P<0.05 versus pGRIM-19 groups). (b) Detection via FCS of apoptosis stimulated by recombinant treatment plasmids, Q1=dead cells, Q2=late apoptotic cells, Q3=normal cells, Q=early apoptotic cells. (c) Representative fluoromicrographs of apoptosis detected by AO/EB assay. The orange labeling indicates early stage apoptotic cells and the red labeling indicates late stage apoptotic cells. (d) Apoptosis of DU145 cells treated with psi-Survivin, pGRIM-19, pGRIM-19-si-Survivin and psi-Scramble plasmids by TUNEL staining, the brown labeling indicates the apoptotic cells (scale bar=20 µm). AO/EB, acridine orange/ethidium bromide; FCS, fetal calf serum; GRIM-19, gene associated with retinoid-interferon-induced mortality-19; RT-PCR, reverse transcription PCR; TUNEL, terminal deoxynucleotidyl transferase-mediated nick end labeling.
To determine if these constructs were able to induce apoptosis in DU145 cells, four different methods were used: flow cytometry, AO/EB staining (nuclear condensation), Annexin V-FITC and TUNEL staining. Flow cytometry showed that in the pGRIM-19-si-Survivin-treated cells, the apoptosis was significantly higher than that in the single-agent-treated cells, psi-Scramble transfection did not significantly affect apoptosis when compared to mock-transfected DU145 (Table 1). In another approach, after 72 h of transfection with the plasmids, cells were stained with Annexin V-FITC to visualize apoptotic cells. Although all groups had a significant increase in apoptosis, compared to the control, pGRIM-19-si-Survivin plasmid was the strongest inducer of apoptosis (Figure 2b). The AO/EB staining revealed that both early apoptotic and late apoptotic DU145 cells were observed in the cells treated with psi-Survivin, pGRIM-19 and pGRIM-19-si-Survivin. In pGRIM-19-si-Survivin group more apoptotic cells were found than those found in single-gene transfectants, which suggested that pGRIM-19-si-Survivin not only had an effect on induction of early apoptosis, but also late apoptosis in DU145 cells (Figure 2c). Consistent with these observations, TUNEL assays also showed the synergistic apoptotic effect of pGRIM-19-si-Survvin (Figure 2d, Supplementary Table 4).
Table 1. Induction of apoptosis by recombinant plasmids and cell cycle analysis in DU145 cells (mean±s.e.).
| Treatment group (n=3) | Apoptotic cells (%) | G0–G1 (%) | S (%) |
|---|---|---|---|
| Mock | 1.89±0.16 | 41.92±1.84 | 16.66±4.66 |
| psi-Survivin | 10.31±1.23*§# | 59.43±2.09*§ | 29.73±3.62*§ |
| pGRIM-19 | 7.94±1.17*§# | 50.22±3.41 | 37.86±4.94*§ |
| pGRIM-19-si-Survivin | 21.83±6.67**§§ | 60.08±4.31*§ | 36.33±3.78*§ |
| psi-Scramble | 3.67±0.96 | 35.73±3.26 | 49.25±2.67 |
*P<0.05, **P<0.01 versus mock group; §P<0.05, §§P<0.01 versus psi-Scramble group; #P<0.05 versus pGRIM-19-si-Survivin.
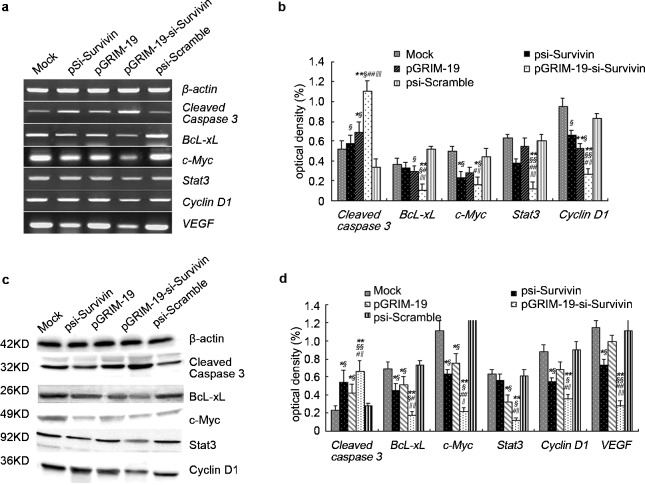
pGRIM-19-si-Survivin co-expression plasmid suppresses BcL-xL, c-Myc, Stat3 and CyclinD1 expression, but increases cleaved caspase3 in DU145 cells
Recent studies indicate that a constitutively active Survivin induces the expression of several genes, including BcL-xL, c-Myc, Stat3, Cyclin D1 and VEGF (vascular endothelial growth factor), which promote cell division. To determine the expression of these gene products, RT-PCR and Western blot analyses were employed. RT-PCR analyses showed that levels of BcL-xL, c-Myc, Stat3, CyclinD1 and VEGF transcripts were significantly decreased in presence of pGRIM-19-si-Survivin, but cleaved caspase3 levels increased compared to plasmids individual gene transfections (Figure 3a and b). Western blots showed an increased expression and cleavage of caspase3 and reduced expression of BcL-xL, c-Myc, Stat3, CyclinD1 and VEGF levels in the presence of pGRIM-19-si-Survivin (Figure 3c and d) when compared to the controls.
Figure 3.
Expression of tumor-associated-genes and proteins in DU145 cells transfected with different treatment plasmids after 48 h. (a) Tumor-associated gene expression in DU145 cells transfected with psi-Survivin, pGRIM-19, pGRIM-19-si-Survivin and psi-Scramble empty vector revealed by RT-PCR. (b) The quantified cleaved caspase3, BcL-xL and c-Myc, Stat3, Cyclin D1 and VEGF gene level in DU145 cells transfected with different vectors. (c) Tumor-associated protein expression in DU145 cells transfected with psi-Survivin, pGRIM-19, pGRIM-19-si-Survivin and psi-Scramble empty vector revealed by Western blot. (d) The quantified cleaved caspase3, BcL-xL, C-Myc, Stat3 and cyclin D1 protein levels in DU145 cells transfected with different treatment vectors. Optical density was expressed as mean±s.e. (*P<0.05, **P<0.01 versus mock; §P<0.05, §§P<0.01 versus psi-Scramble group; #P<0.05, # # P<0.01 versus psi-Survivin; ‖‖P<0.05, ‖‖‖‖P<0.01 versus pGRIM-19 group). BcL-XL, B-cell lymphoma xL; GRIM-19, gene associated with retinoid-interferon-induced mortality-19; RT-PCR, reverse transcription PCR; Stat3, signal transducer and activator of transcription 3; VFGF, vascular endothelial growth factor.
Bacteria-mediated transfer of pGRIM-19-si-Survivin in vivo synergistically suppresses tumor growth
We next employed a xenograft model to study the impact of pGRIM-19-si-Survivin on tumor growth. Since systemically introduced plasmids do not reach tumors in optimal concentrations to ablate growth, we employed S. typhimurium as a carrier for the plasmids under study. Although Salmonella has been reported to retard the growth of established tumors significantly by itself, tumor regression is incomplete. Mice were transplanted with DU145 cells subcutaneously. Once palpable tumors have developed, mice were divided into six groups (n=5 per group) and then injected intraperitoneally with 1×107 CFU of attenuated Salmonella carrying different plasmids. Tumor growth was monitored weekly (Figure 4a). On day 70, mice were killed and final tumor weights and volumes were determined (Table 2). As shown in Table 2, mice treated with buffer alone (mock control) developed tumors with a mean volume of 4897.41±1289.37 mm3. In mice treated with attenuated S. typhimurium only, tumors grew to a volume of 4260.22±1303.11 mm3; in mice treated with attenuated Salmonella carrying psi-Scramble plasmid, the tumor grew to 4528.09±735.21 mm3. Thus, the bacteria carrying the Scramble-shRNA did not significantly affect tumor growth compared to Salmonella alone. However, mice treated with Salmonella-psi-Survivin, Salmonella-pGRIM-19 and Salmonella-pGRIM-19-si-Survivin developed tumors with a median reduced volume of 671.36±165.11 mm3, 857.29±170.07 mm3 and 284.33±57.33 mm3, respectively. Although Salmonella appeared to inhibit tumor growth slightly, no significant differences were observed in tumor volumes between buffer control and Salmonella treatment alone. The differences between Salmonella-psi-Scramble or Salmonella alone versus Salmonella-psi-Survivin, Salmonella-pGRIM-19 and Salmonella-pGRIM-19-si-Survivin groups were highly significant (P<0.01). The differences between Salmonella-psi-Survivin, Salmonella-pGRIM-19 and Salmonella-pGRIM-19-si-Survivin groups in tumor size were also statistically significant. In summary, compared with Salmonella-psi-Scramble group, seven-fold in Salmonella-psi-Survivin, 5.5-fold in Salmonella-pGRIM-19 and 16-fold in Salmonella-pGRIM-19-si-Survivin higher tumor suppressive effects were achieved respectively. Although S. typhimurium carrying psi-Survivin or pGRIM-19 show significant antitumor effect, it can be further enhanced by co-expression of both gene products.
Figure 4.
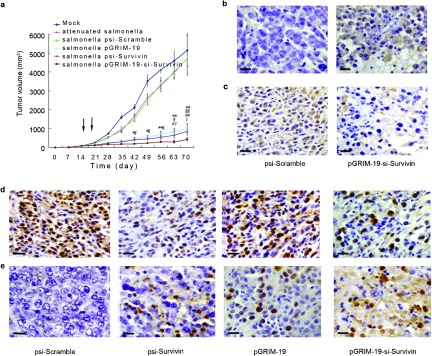
Intraperitoneal injection of attenuated Salmonella carrying different treatment plasmids resulted in significant inhibition of tumor growth and induction of apoptosis in prostate cancer grafts in vivo. (a) Growth curves of DU145 xenografts treated with attenuated Salmonella carrying recombinant plasmids (*P<0.05, **P<0.01 versus mock, §P<0.05, §§P<0.01 versus psi-Scramble group; #P<0.05, # # P<0.01versus psi-Survivin; ‖‖P<0.05, versus pGRIM-19 group). Arrows show Salmonella treatment carrying different plasmids through intraperitoneal at days 14 and 21. (b) Immunohistochemical analysis of Survivin expression for DU145 xenografts treated with S. typhimurium carrying psi-Scramble and pGRIM-19-si-Survivin plasmids. (c) Immunohistochemical analysis for GRIM-19 expression in DU145 xenografts treated with S. typhimurium carrying psi-Scramble and pGRIM-19-si-Survivin plasmids. (d) Immunohistochemical staining for Ki-67 in DU145 in vivo grafts; positive cells are brown. (e) TUNEL assay; TUNEL-positive cells are brown (scale bar=20 µm). GRIM-19, gene associated with retinoid-interferon-induced mortality-19; S. typhimurium, Salmonella enterica serovar typhimurium; TUNEL, terminal deoxynucleotidyl transferase-mediated nick end labeling.
Table 2. Comparison of the mean weight of nude mice, and mean weight/volume of tumors after different treatments (mean±s.e.).
| Mean weight of nude mice (g) | Mean weight of tumor (g) | Mean volume of tumor (mm3) | |
|---|---|---|---|
| Mock | 16.32±2.06 | 6.72±1.53 | 4897.41±1289.37 |
| Attenuated Salmonella alone | 18.33±4.61 | 5.73±1.62 | 4260.22±1303.11 |
| Salmonella -psi-Scramble | 15.76±2.63 | 5.92±1.39 | 4528.09±735.21 |
| Salmonella -pGRIM-19 | 24.62±3.36* | 2.62±0.75*# | 857.29±170.07*# |
| Salmonella- psi-Survivin | 25.12±3.77* | 2.04±0.71*# | 671.36±165.11*# |
| Salmonella -pGRIM-19-si-Survivin | 27.32±2.98* | 1.26±0.42** | 284.33±57.33** |
P<0.05 versus Salmonella-pGRIM-19-si-Survivin; *P<0.05, **P<0.01 versus corresponding Salmonella-psi-Scramble.
Apart from measuring the tumor volumes, we employed IHC, H&E staining and TUNEL assays to demonstrate the antitumor activities. IHC showed that Survivin expression was inhibited (Figure 4b), while that of GRIM-19 was enhanced (Figure 4c). Consistent with the antitumor properties Ki-67 expression was repressed (Figure 4d, Supplementary Table 4). TUNEL staining showed a massive apoptosis, necrosis in Salmonella-pGRIM-19-si-Survivin-treated tumors. In sharp contrast to this, the Scramble shRNA control showed high Ki-67-positive cells and little TUNEL-positive cells (Figure 4e).
pGRIM-19-si-Survivin co-expression promotes DU145 cells apoptosis in vivo
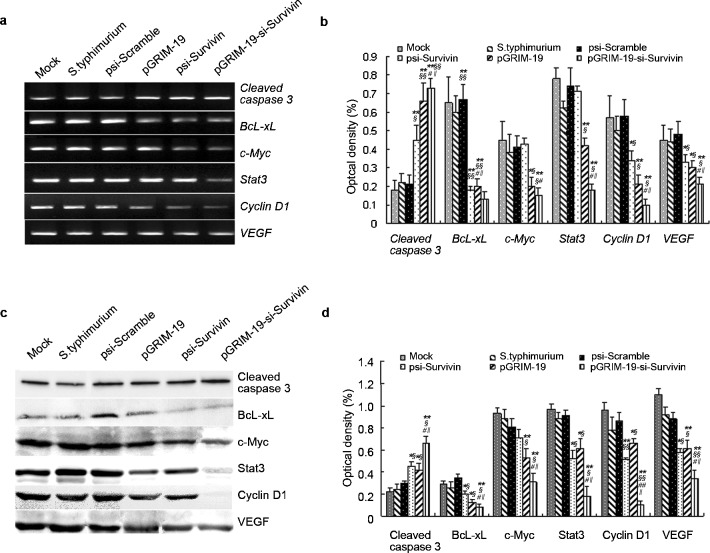
To further demonstrate the effect of pGRIM-19-si-Survivin on tumors, we measured gene expression using RT-PCR and protein abundance using Western blot analysis. Overexpression of Survivin and little or no expression GRIM-19 were observed in the control tumors. In pGRIM-19-si-Survivin group expression of Survivin was significantly inhibited and that of GRIM-19 was enhanced (Supplementary Figure 2). We also measured apoptosis associated gene expression in these tumors. The results indicated that BcL-xL, c-Myc, Stat3, CyclinD1 and VEGF mRNA were significantly decreased in group treated with pGRIM-19-si-Survivin, while cleaved caspase3 expression was elevated significantly (Figure 5a and b). Western blot analysis also showed that the cleaved caspase3 level was significantly increased; while those of BcL-xL, c-Myc and VEGF were decreased in the pGRIM-19-si-Survivin group (P<0.05 versus the control groups) and Stat3, CyclinD1 were extremely decreased in this group (P<0.01 versus the control groups) (Figure 5c and d).
Figure 5.
Expression of tumor-associated genes and proteins in DU145 prostate cancer grafts treated with attenuated Salmonella carrying different treatment plasmids. (a) RT-PCR analyses for cleaved capase3, BcL-xL, c-Myc, Stat3, Cyclin D1 and VEGF expression in DU145 prostate cancer graft with attenuated Salmonella carrying different treatment plasmids. (b) Quantified mRNA levels in xenografts treated with recombinnant bacteria carrying various plasmids. (c) Western blot analyses for cleaved Capase3, BcL-xL, c-Myc, Stat3, Cyclin D1 and VEGF expression in DU145 prostate cancer grafts treated with attenuated Salmonella carrying different treatment plasmids; (d) Quantified protein levels in xenografts treated with recombinnant bacteria carrying various plasmids. Mean±s.e. (**P<0.01 versus mock; §P<0.05, §§P<0.01 versus psi-Scramble group; #P<0.05, # # P<0.01 versus psi-Survivin; ‖‖P<0.05 versus pGRIM-19 group). BcL-XL, B-cell lymphoma xL; GRIM-19, gene associated with retinoid-interferon-induced mortality-19; RT-PCR, reverse transcription PCR; Stat3, signal transducer and activator of transcription 3; VEGF, vascular endothelial growth factor.
Discussion
Current treatments for advanced androgen-independent PCa are only marginally effective. Thus, it is important to understand the molecular mechanisms involved in the pathogenesis and progression of this disease to identify novel therapeutic targets and develop effective treatment strategies. Elevated Survivin expression has been detected in primary tissues and cell lines of prostate tumors.33 The accumulating data indicate that Survivin activates several genes whose products promote cell cycle progression (e.g., Cyclin D1, c-Myc and Stat3), prevent apoptosis (e.g., caspase3, Bcl-XL), or promote vascular formation (e.g., VEGF).32, 34, 35, 36, 37
In this study, we demonstrated that Survivin and GRIM-19 play a key role in prostate tumor growth. Immunohistochemical, Western blot and RT-PCR analysis showed that Survivin was overexpressed both in human PCa DU145 cells and human primary PCa tissues. These data are consistent with other studies5, 8, 15 that showed an abnormal activation of Survivin in PCa. The importance of GRIM-19 in tumor growth regulation has been realized recently. GRIM-19 was isolated as an inhibitor of cell growth in response to IFN–retinoid treatment. It has been shown to interact exclusively with the transcriptional activating domain of Stat3 and inhibited transcriptional activation.27, 28, 38 Recent studies have also shown that GRIM-19 expression is reduced or eliminated in a number of primary tumors.29, 35 Survivin is a downstream target of Stat3. Thus, Survivin and GRIM-19 have become promising targets for cancer therapy.3, 39
The use of RNAi against Survivin produced antitumor effects both in vitro and in vivo, and although suppression of tumor growth was significant, it was incomplete. Many factors, such as the RNA sequence, expression levels and stability, determine the efficacy of shRNAs. In addition, the rate of transcription of the target gene can affect the extent of target mRNA destruction by shRNAs and therefore the efficacy of this approach. However, the moderate antitumor effects of Survivin-shRNA could be strengthened by adding another inhibitor. In this study, we have demonstrated the superior antitumor activity of Survivin-specific shRNAs when combined with GRIM-19 co-expressed from the same plasmid.
Both Cyclin D1 and c-Myc respond to mitogenic signals and are required for progression of cells from G1 to S phase of the cell cycle.40 BcL-xL is one of the important regulators of apoptosis and is involved in the regulation of the cell cycle, prolongs cell survival and inhibits apoptosis. Stat3 normally resides in the cytoplasm and can be activated through phosphorylation by cytokines, growth factors and oncogenic proteins. Activated Stat3 forms homodimers and translocates to the nucleus, where its occupation of specific DNA-binding sites results in the increased transcription of several molecules that are involved directly in cell survival and proliferation. Stat3 is downstream of GRIM-19, so CyclinD1, BcL-xL and c-Myc can be regulated by Survivin. Our data showed that the expression of Cyclin D1, BcL-xL, Stat3 and c-Myc was synergistically diminished in cells transfected with pGRIM-19-si-Survivin. However, cleaved caspase3 expression was synergistically upregulated under these conditions. These results demonstrated that ablation of Survivin signaling and increasing GRIM-19 at the same time was accompanied by growth inhibition and induction of apoptosis in vitro. Thus, inhibition of tumor growth under these circumstances appears to be due both to a combination of cell cycle arrest and to activation of apoptosis.
Chemical synthesis of small interfering RNAs (siRNAs) is not cost-effective for large scale therapeutic applications. RNAi caused by synthetic siRNAs is transient due to lack of tumor-targeting and degradation of siRNA.40 However, this problem has been solved here by using plasmid expression vectors in Salmonella as a delivery tool. These plasmid vectors produce sufficient amounts of shRNA using normal cellular machinery,41 and are able to knockdown expression of target genes for extended periods of time.
The success of shRNAs as cancer therapeutics relies on the development of safe, economical and efficacious in vivo delivery systems into tumor cells in vivo. Although shRNAs can be used as therapeutics in vivo, their intratumoral delivery, specifically across the plasma membrane of cells is not achieved easily. Viral delivery vectors possess low replication packaging efficiency (i.e., small capacity for encoding extra sequences) and do not target tumors. Nanoparticles, though resisting degradation, have poor targeting and low potency, so their clinical application is yet limited. The most ideal delivery system would be: (i) nontoxic to normal cells; and (ii) able to deliver the therapeutic efficiently and specifically to the tumor. S. typhimurium possesses many qualities that make it amenable for potential use in humans: data from mice showing tumor targeting ability and tumor inhibition, genetic stability and antibiotic sensitivity. A barrier to utilizing bacteria safely as systemically administered anticancer agents in humans is that bacteria often over stimulate TNF-α induction, which leads to a cytokine cascade responsible for septic shock in humans, typically seen during systemic bacterial infections.42 In Gram-negative bacteria, this potentially lethal effect is due to the response generated by myristoylation of lipid A in the bacterial lipopolysaccharide. The msbB gene of Salmonellae is responsible for the terminal myristoylation of lipid A.43, 44 Disruption of the msbB gene in Salmonellae results in stable mutants that lower TNF induction. Introducing msbB2 mutations into Salmonella strain YS72 (hyperinvasive, purI2, xyl2), previously used for tumor targeting, has demonstrated that introduction of these mutations does not interfere with the ability of the Salmonellae to target tumors.45 S. typhimurium strain VNP20009 contains deletions in purI and msbB, generating a genetically stable, antibiotic-sensitive strain that has dramatically reduced TNF-α induction in mice and pigs while retaining tumor-targeting and tumor-inhibition properties.
Since hypoxia is a common characteristic of human solid tumors, which adversely affects the prognosis of cancer patients, targeting the hypoxic regions of tumors may increase the effectiveness of cancer treatment. Salmonella can accumulate in hypoxic tumor cells. Thus, anti-tumor genes vectored by Salmonella can be functionally transferred to mammalian cells for anti-tumor therapy.46 The resulting nutritional requirements of Salmonella were apparently met within the tumor environment where the bacteria replicated/concentrated to at least 1000-fold over their level in normal tissues. Preferential replication allows the bacteria to produce and deliver a variety of anticancer therapeutic agents at high concentrations directly within the tumor, while minimizing toxicity to normal tissues.47, 48 Attenuated S. typhimurium has been found to be safe in mice, pigs and monkeys when administered intravenously or by intra-abdominal routes. Thus, the combination of live attenuated bacteria as carriers for shRNAs and tumor growth inhibitors holds strong promise for tumor suppression.
Herein, we examined the inhibitory effects of a combined GRIM-19/Survivin-specific shRNA carried by attenuated S. typhimurium on tumor growth, and found it was superior to either component alone. Indeed, the combined therapy completely inhibited the activity of Ki-67 and VEGF and also downregulated the expression of Cyclin D1 and c-Myc, BcL-xL and Stat3. The relatively high targeting specificity of attenuated S. typhimurium plus the different mechanisms of Survivin downregulation are key properties that contribute to the observed synergistic antitumor effects of the combination vector. Further efforts will clearly be necessary to explore the human therapeutic value of using these recombinant plasmids together with attenuated Salmonella as a carrier and in multiple doses to totally eliminate carcinomas. Employing an appropriate conventional antitumor treatment combined with targeted genetic therapies (e.g., siRNA) may ultimately achieve an improved therapeutic effect while allowing for lowered doses for each component, perhaps reducing total adverse effects.
Most importantly, our data showed that inhibition of Survivin by administration of appropriate vector-based shRNAs and co-expression of GRIM-19 carried by attenuated Salmonella was an effective and feasible approach to PCa therapy through intraperitoneal injection. The feasibility of this approach to treat tumors clinically has not been fully explored, although it seems logical. In fact, attenuated Salmonella vectors have already been applied in vivo for the treatment of some malignancies, such as PCa.24 Further efforts are clearly necessary for exploring the therapeutic value of parenteral administration of attenuated Salmonella carrying various anti-tumor therapies. In addition, the use of additional cell lines would be informative. In summary, we present evidence for the potential use of shRNAs for blocking persistent Survivin, in association with GRIM-19 to reverse tumor growth. Bacteria-delivered plasmid-based shRNA therapy for tumor suppression may be an effective, inexpensive approach and delivery system for cancer therapy. Future studies will aim at using Salmonella vectors or other tumor-targeting delivery systems containing combined antitumor therapies to evaluate the full potential of a multifactorial antitumor approach.
Author contributions
YBL carried out the molecular and animal studies, LZ participated in its design of the study and drafted the manuscript, YXG carried out the animal studies, LFG participated in its design of the study, XCL performed the statistical analysis, LJZ carried out the molecular studies, BFG carried out the animal studies, LJZ coordinated and helped to draft the manuscript, XJZ participated in its design of the study and DQX conceived of the study, and helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 30801354 and No. 3097079J), Jilin Provincial Science and Technology Department (No. 20080154 and No.20110146) and the PhD Programs Foundation of Ministry of Education of China (No. 200801831077) (Ling Zhang).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Asian Journal of Andrology's website (http://www.nature.com/aja).
Supplementary Information
References
- Pisani P, Parkin DM, Ferlay J. Estimates of the worldwide mortality from eighteen major cancers in 1985. Implications for prevention and projections of future burden. Int J Cancer. 1993;55:891–903. doi: 10.1002/ijc.2910550604. [DOI] [PubMed] [Google Scholar]
- Nicholson B, Theodorescu D. Molecular therapeutics in prostate cancer. Histol Histopathol. 2003;18:275–98. doi: 10.14670/HH-18.275. [DOI] [PubMed] [Google Scholar]
- Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22:8581–9. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, Survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–21. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Madden SL, Zhang L, Lash AE, Yu J, et al. Analysis of human ranscriptomes. Nat Genet. 1999;23:387–8. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- Li F, Ling X. Survivin study: an update of ‘what is the next wave'. J Cell Physiol. 2006;208:476–86. doi: 10.1002/jcp.20634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman KW, Li Y, Wang Z, Sarkar SH, Sarkar FH. Gene expression profiling revealed Survivin as a target of 3,3′-diindolylmethane-induced cell growth inhibition and apoptosis in breast cancer cells. Cancer Res. 2006;66:4952–60. doi: 10.1158/0008-5472.CAN-05-3918. [DOI] [PubMed] [Google Scholar]
- Li F, Ackermann EJ, Bennett CF, Rothermel AL, Plescia J, et al. Pleiotropic cell-division defects and apoptosis induced by interference with Survivin function. Nat Cell Biol. 1999;1:461–6. doi: 10.1038/70242. [DOI] [PubMed] [Google Scholar]
- Ling X, Li F. Silencing of antiapoptotic Survivin gene by multiple approaches of RNA interference technology. Biotechniques. 2004;36:450–4,–6–60. doi: 10.2144/04363RR01. [DOI] [PubMed] [Google Scholar]
- Tu SP, Jiang XH, Lin MC, Cui JT, Yang Y, et al. Suppression of Survivin expression inhibits in vivo tumorigenicity and angiogenesis in gastric cancer. Cancer Res. 2003;63:7724–32. [PubMed] [Google Scholar]
- Choi KS, Lee TH, Jung MH. Ribozyme-mediated cleavage of the human Survivin mRNA and inhibition of antiapoptotic function of Survivin in MCF-7 cells. Cancer Gene Ther. 2003;10:87–95. doi: 10.1038/sj.cgt.7700531. [DOI] [PubMed] [Google Scholar]
- Wall NR, O'Connor DS, Plescia J, Pommier Y, Altieri DC. Suppression of Survivin phosphorylation on Thr34 by flavopiridol enhances tumor cell apoptosis. Cancer Res. 2003;63:230–5. [PubMed] [Google Scholar]
- Zhang M, Yang J, Li F. Transcriptional and post-transcriptional controls of Survivin in cancer cells: novel approaches for cancer treatment. J Exp Clin Cancer Res. 2006;25:391–402. [PMC free article] [PubMed] [Google Scholar]
- Hayashi N, Asano K, Suzuki H, Yamamoto T, Tanigawa N, Egawa S, et al. Adenoviral infection of Survivin antisense sensitizes prostate cancer cells to etoposide in vivo. . Prostate. 2005;65:10–9. doi: 10.1002/pros.20232. [DOI] [PubMed] [Google Scholar]
- Pennati M, Binda M, Colella G, Zoppe M, Folini M, et al. Ribozyme-mediated inhibition of Survivin expression increases spontaneous and drug-induced apoptosis and decreases the morigenic potential of human prostate cancer cells. Oncogene. 2004;23:386–94. doi: 10.1038/sj.onc.1207071. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhao L, Zhao D, Lin G, Guo B, et al. Inhibition of tumor growth and induction of apoptosis in prostate cancer cell lines by overexpression of tissue inhibitor of matrix metalloproteinase-3. Cancer Gene Ther. 2010;17:171–9. doi: 10.1038/cgt.2009.59. [DOI] [PubMed] [Google Scholar]
- Maximo V, Botelho T, Capela J, Soares P, Lima J, et al. Somatic and germline mutation in GRIM-19, a dual function gene involved in mitochondrial metabolism and cell death, is linked to mitochondrion-rich (Hurthle cell) tumours of the thyroid. Br J Cancer. 2005;92:1892–8. doi: 10.1038/sj.bjc.6602547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alchanati I, Nallar SC, Sun P, Gao L, Hu J, et al. A proteomic analysis reveals the loss of expression of the cell death regulatory gene GRIM-19 in human renal cell carcinomas. Oncogene. 2006;25:7138–47. doi: 10.1038/sj.onc.1209708. [DOI] [PubMed] [Google Scholar]
- Glienke W, Maute L, Wicht J, Bergmann L. Curcumin inhibits constitutive STAT3 phosphorylation in human pancreatic cancer cell lines and downregulation of survivin/BIRC5 gene expression. Cancer Invest. 2010;28:166–71. doi: 10.3109/07357900903287006. [DOI] [PubMed] [Google Scholar]
- Ashizawa T, Miyata H, Ishii H, Oshita C, Matsuno K, et al. Antitumor activity of a novel small molecule STAT3 inhibitor against a human lymphoma cell line with high STAT3 activation. Int J Oncol. 2011;38:1245–52. doi: 10.3892/ijo.2011.957. [DOI] [PubMed] [Google Scholar]
- Sun M, Liu C, Nadiminty N, Lou W, Zhu Y, et al. Inhibition of Stat3 activation by sanguinarine suppresses prostate cancer cell growth and invasion. Prostate. 2012;72:82–9. doi: 10.1002/pros.21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts TR, DeVico AL, Onyabe DY, Shata MT, Bagley KC, et al. Progress toward the development of a bacterial vaccine vector that induces high-titer long-lived broadly neutralizing antibodies against HIV-1. FEMS Immunol Med Microbiol. 2003;37:129–34. doi: 10.1016/S0928-8244(03)00067-1. [DOI] [PubMed] [Google Scholar]
- Nguyen VH, Kim HS, Ha JM, Hong Y, Choy HE, et al. Genetically engineered Salmonella typhimurium as an imageable therapeutic probe for cancer. Cancer Res. 2010;70:18–23. doi: 10.1158/0008-5472.CAN-09-3453. [DOI] [PubMed] [Google Scholar]
- Zhang L, Gao L, Zhao L, Guo B, Ji K, et al. Intratumoral delivery and suppression of prostate tumor growth by attenuated Salmonella enterica serovar typhimurium carrying plasmid-based small interfering RNAs. Cancer Res. 2007;67:5859–64. doi: 10.1158/0008-5472.CAN-07-0098. [DOI] [PubMed] [Google Scholar]
- Ning JF, Zhu W, Xu JP, Zheng CY, Meng XL. Oral delivery of DNA vaccine encoding VP28 against white spot syndrome virus in crayfish by attenuated Salmonella typhimurium. . Vaccine. 2009;27:1127–35. doi: 10.1016/j.vaccine.2008.11.075. [DOI] [PubMed] [Google Scholar]
- Xu DQ, Zhang L, Kopecko DJ, Gao L, Shao Y, et al. Bacterial delivery of siRNAs: a new approach to solid tumor therapy. Methods Mol Biol. 2009;487:161–87. doi: 10.1007/978-1-60327-547-7_8. [DOI] [PubMed] [Google Scholar]
- Hu J, Angell JE, Zhang J, Ma X, Seo T, et al. Characterization of monoclonal antibodies against GRIM-19, a novel IFN-beta and retinoic acid-activated regulator of cell death. J Interferon Cytokine Res. 2002;22:1017–26. doi: 10.1089/107999002760624242. [DOI] [PubMed] [Google Scholar]
- Gao L, Zhang L, Hu J, Li F, Shao Y, et al. Down-regulation of signal transducer and activator of transcription 3 expression using vector-based small interfering RNAs suppresses growth of human prostate tumor in vivo. . Clin Cancer Res. 2005;11:6333–41. doi: 10.1158/1078-0432.CCR-05-0148. [DOI] [PubMed] [Google Scholar]
- Zhang L, Gao L, Li Y, Lin G, Shao Y, et al. Effects of plasmid-based Stat3-specific short hairpin RNA and GRIM-19 on PCa-3M tumor cell growth. Clin Cancer Res. 2008;14:559–68. doi: 10.1158/1078-0432.CCR-07-1176. [DOI] [PubMed] [Google Scholar]
- Hohmann EL, Oletta CA, Killeen KP, Miller SI. phoP/phoQ-deleted Salmonella typhi (Ty800) is a safe and immunogenic single-dose typhoid fever vaccine in volunteers. J Infect Dis. 1996;173:1408–14. doi: 10.1093/infdis/173.6.1408. [DOI] [PubMed] [Google Scholar]
- Chen YZ, Li ZD, Gao F, Zhang Y, Sun H, et al. Effects of combined Chinese drugs and chemotherapy in treating advanced non-small cell lung cancer. Chin J Integr Med. 2009;15:415–9. doi: 10.1007/s11655-009-0415-2. [DOI] [PubMed] [Google Scholar]
- Weng JR, Tsai CH, Kulp SK, Wang D, Lin CH, et al. A potent indole-3-carbinol derived antitumor agent with pleiotropic effects on multiple signaling pathways in prostate cancer cells. Cancer Res. 2007;67:7815–24. doi: 10.1158/0008-5472.CAN-07-0794. [DOI] [PubMed] [Google Scholar]
- Gong J, Lee J, Akio H, Schlegel PN, Shen R. Attenuation of apoptosis by chromogranin A-induced Akt and survivin pathways in prostate cancer cells. Endocrinology. 2007;148:4489–99. doi: 10.1210/en.2006-1748. [DOI] [PubMed] [Google Scholar]
- Cosgrave N, Hill AD, Young LS. Growth factor-dependent regulation of Survivin by c-Myc in human breast cancer. J Mol Endocrinol. 2006;37:377–90. doi: 10.1677/jme.1.02118. [DOI] [PubMed] [Google Scholar]
- Wang R, Lin F, Wang X, Gao P, Dong K, et al. Suppression of Bcl-xL expression by a novel tumor-specific RNA interference system inhibits proliferation and enhances radiosensitivity in prostatic carcinoma cells. Cancer Chemother Pharmacol. 2008;61:943–52. doi: 10.1007/s00280-007-0548-y. [DOI] [PubMed] [Google Scholar]
- Scheper MA, Nikitakis NG, Sauk JJ. Survivin is a downstream target and effector of sulindac-sensitive oncogenic Stat3 signalling in head and neck cancer. Int J Oral Maxillofac Surg. 2007;36:632–9. doi: 10.1016/j.ijom.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Singh RP, Deep G, Blouin MJ, Pollak MN, Agarwal R. Silibinin suppresses in vivo growth of human prostate carcinoma -3 tumor xenograft. Carcinogenesis. 2007;28:2567–74. doi: 10.1093/carcin/bgm218. [DOI] [PubMed] [Google Scholar]
- Liang ZW, Guo BF, Li Y, Li XJ, Li X, et al. Plasmid-based Stat3 siRNA delivered by hydroxyapatite nanoparticles suppresses mouse prostate tumour growth in vivo. Asian J Androl. 2011;13:481–6. doi: 10.1038/aja.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani P, Parkin DM, Bray F, Ferlay J. Estimates of the worldwide mortality from 25 cancers in 1990. Int J Cancer. 1999;83:18–29. doi: 10.1002/(sici)1097-0215(19990924)83:1<18::aid-ijc5>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Dechow TN, Pedranzini L, Leitch A, Leslie K, Gerald WL, et al. Requirement of matrix metalloproteinase-9 for the transformation of human mammary epithelial cells by Stat3-C. Proc Natl Acad Sci USA. 2004;101:10602–7. doi: 10.1073/pnas.0404100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Weber K, Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- Carswell EA, Old LJ, Kassel RL, Green S, Fiore N, et al. An endotoxin-induced serum factor that causes necrosis of tumors. Proc Natl Acad Sci USA. 1975;72:3666–70. doi: 10.1073/pnas.72.9.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SA, Everest P, Servos S, Foxwell N, Zahringer U, et al. A lethal role for lipid A in Salmonella infections. Mol Microbiol. 1998;29:571–9. doi: 10.1046/j.1365-2958.1998.00952.x. [DOI] [PubMed] [Google Scholar]
- Clementz T, Zhou Z, Raetz CR. Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A. Acylation by MsbB follows laurate incorporation by HtrB. J Biol Chem. 1997;272:10353–60. doi: 10.1074/jbc.272.16.10353. [DOI] [PubMed] [Google Scholar]
- Low KB, Ittensohn M, Le T, Platt J, Sodi S, et al. Lipid A mutant Salmonella with suppressed virulence and TNFalpha induction retain tumor-targeting in vivo. Nat Biotechnol. 1999;17:37–41. doi: 10.1038/5205. [DOI] [PubMed] [Google Scholar]
- Clairmont C, Lee KC, Pike J, Ittensohn M, Low KB, et al. Biodistribution and genetic stability of the novel antitumor agent VNP20009, a genetically modified strain of Salmonella typhimurium. J Infect Dis. 2000;181:1996–2002. doi: 10.1086/315497. [DOI] [PubMed] [Google Scholar]
- Zhao M, Yang M, Li XM, Jiang P, Baranov E, et al. Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proc Natl Acad Sci USA. 2005;102:755–60. doi: 10.1073/pnas.0408422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Yang M, Ma H, Li X, Tan X, et al. Targeted therapy with a Salmonella typhimurium leucine-arginine auxotroph cures orthotopic human breast tumors in nude mice. Cancer Res. 2006;6:7647–52. doi: 10.1158/0008-5472.CAN-06-0716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.