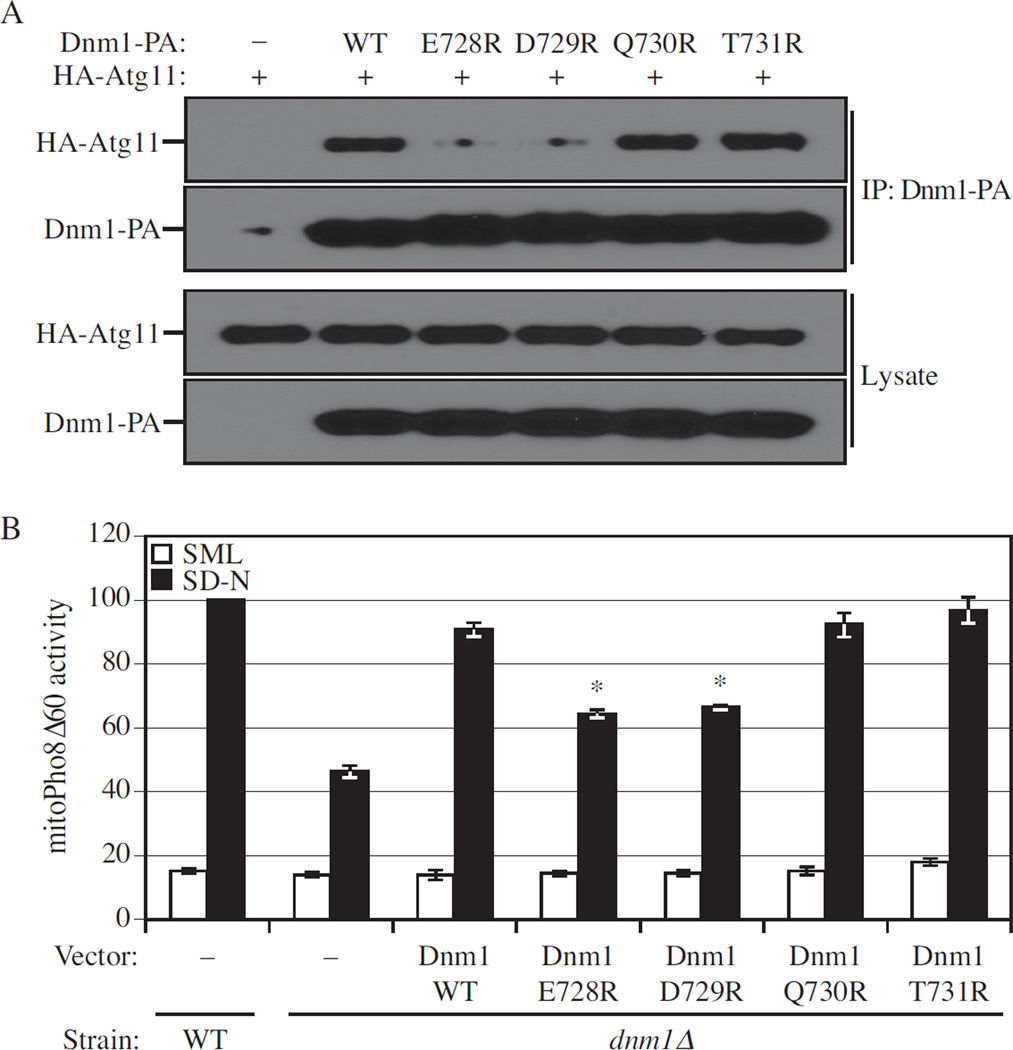
Figure 7. Dnm1 Mutants that Lose Binding to Atg11 are Mitophagy Defective.
(A) The plasmid pCuHA-Atg11 together with pDnm1-PA, pDnm1(E728R)-PA, pDnm1(D729R)-PA, pDnm1(Q730R)-PA, or pDnm1(T731R)-PA was co-transformed into atg11 Δ dnm1 Δ (KDM1251) cells. Cells were cultured in SML and shifted to SD-N for 1.5 h. Cell lysates were prepared and incubated with IgG-Sepharose for affinity isolation as described in Experimental Procedures. The eluted proteins were separated by SDS-PAGE and detected with monoclonal anti-HA antibody and an antibody that binds to PA.
(B) MitoPho8Δ60 wild-type (KDM1009) cells were transformed with empty vector; mitoPho8Δ60 dnm1 Δ (KDM1014) cells were transformed with empty vector, pDnm1-PA, pDnm1(E728R)-PA, pDnm1(D729R)-PA, pDnm1(Q730R)-PA, or pDnm1(T731R)-PA. Cells were cultured in SML to mid-log phase, then shifted to SD-N for 6 h. The mitoPho8Δ60 assay was performed as described in Experimental Procedures. Error bars correspond to the standard error, and were obtained from three independent repeats. * p<0.01.
Also see Fig. S4.