Abstract
There is increasing evidence that the high mobility group A1 (HMGA1) protein, which functions as a transcriptional master regulator, plays critical roles in tumor progression. We evaluated HMGA1 expression in 89 primary uveal melanomas (UM) by immunohistochemistry to determine the clinicopathological and prognostic value of HMGA1 in UM after adjusting for other prognostic variables. Nuclear expression of HMGA1 was detected in 44% UMs. High expression levels of HMGA1 were more frequent in UMs with high levels of epithelioid cell pattern, mitoses count, and Ki67 labeling index (P = 0.025, P<0.0001, P = 0.0018; respectively), and HMGA1 expression levels were directly correlated with Ki67 labeling indexes and mitoses counts (R = 0.31, P <0.0001; R = 0.27, P<0.0068; respectively). High expression of HMGA1 was also independently associated with an increased risk of distant metastases as determined using the Cox proportional hazards regression model (multivariate hazard ratio: 3.44; 95% confidence interval: 1.56–7.60; log rank P = 0.0022). Moreover, high HMGA1 expression was associated with shorter UM-specific survival (multivariate hazard ratio: 2.41; 95% confidence interval: 1.10–5.53; log rank P = 0.041). These findings suggest that high levels of HMGA1 are associated with adverse clinical outcomes in UM patients and that further evaluation of HMGA1 as a potential therapeutic target in UM is warranted.
Introduction
Uveal melanomas (UMs), which arise from neural crest-derived melanocytes within the choroidal plexus of the eye including iris, ciliary body, and choroid are the most common primary intraocular malignant tumors in adults and the most common noncutaneous melanomas. Although they comprise approximately 5% of all diagnosed melanoma cases, their incidence is only 6 per 1 million per year among white and even less among non-white races [1]–[3]. Nevertheless, UMs account for 13% of all deaths from melanoma [4]. In fact, despite the increased diagnostic accuracy and the development of conservative and effective early local treatments of primary intraocular tumors including plaque radiotherapy and photon beam therapy, up to 50% of UM patients die within 15 years of diagnosis from metastases, which almost exclusively involve the liver [3], [5], [6]. Once metastases are diagnosed, the patients' survival time averages no more than 5 to 8 months [7], [8]. presumably due to silent hematogenous systemic micrometastases that occur prior to the diagnosis of clinically evident ocular disease [9], [10]. As no effective therapies are currently available to decrease the risk of metastases, efforts have been made to identify high-risk patients in order to target systemic adjuvant therapy aimed at preventing or delaying metastatic disease.
Many clinicopathological factors including largest basal tumor diameter, age, the presence of epithelioid melanoma cells, closed vascular loops, mitotic activity, nodular growth and extracellular matrix patterns [11]–[15] have been found to be associated with increased risk of developing metastatic disease in UM. Regarding genetic evaluation, chromosome 3 monosomy, mutations of GNAQ or GNA11 and breast cancer 1-associated protein (BAP1) have all been found to impart poor prognosis [16]–[19]. However, none of the above parameters or genes can be considered as therapeutic targets in UM.
High mobility group (HMG) proteins interact with other proteins to alter chromatin structure and so affect gene expression. They are involved in diverse cellular processes, including cell cycle progression, embryologic development, neoplastic transformation, differentiation, apoptosis, cellular metabolism, and DNA repair. In most differentiated tissues of adults, however, they are absent or accumulate to only very low levels [20], [21]. Nevertheless, increasing evidence suggests that a member of this family of proteins, HMGA1, plays a critical role in tumor progression in diverse malignancies. Originally, HMGA1 was described to function as a potent oncogene in cultured cells [22]–[24] and transgenic mice [25], [26]. The expression level of HMGA1 protein was found to be linked to the highly malignant phenotype of human cancers and a poor prognostic indicator, for instance for retinoblastomas [27]–[29]. It has been suggested that HMGA1 might serve not only as a potential biomarker but also as a therapeutic target for advanced malignancies [27]–[29] and metastatic progression [30], [31]. Hence, we speculated that HMGA1 may also play a key role in UM prognosis and as potential target for therapeutic intervention in UM. MIB-1 LI recognizes the nuclear antigen (Ki-67), which makes it an excellent marker of cells in the proliferative phase. In addition to being widely used to assess tumor proliferation rate, MIB-1 LI has prognostic value [32]. In our previous study [28], high MIB-1 LI expression levels were associated with poor prognosis in retinoblastomas, and the increased expression of HMGA1 and HMGA2 correlated with the MIB-1 LI.
In the present study, we explored the prognostic value of HMGA1 by an immunohistochemical analysis in a series of UM cases. The data were then correlated with the relevant clinical information and with the labeling index for the cell proliferation marker Ki67/MIB-1. Furthermore, we discuss the potential of HMGA1 protein as a prognostic marker of UM progression and possible approaches for inhibition of HMGA activity in UM.
Materials and Methods
Ethics statement
This study conformed to the Declaration of Helsinki and was approved by the ethics committee of Qilu Hospital of Shandong Unversity or Beijing Tongren Hospital. All patients had previously given their written informed consent for experimental research on residual tumor tissues available after histopathologic and cytogenetic analyses. The consent procedure was approved by the ethics committee of Qilu Hospital of Shandong Unversity or Beijing Tongren Hospital.
Patients and tumor samples
Eighty-nine primary UM lesions were obtained after enucleation with complete clinical data between 1998 and 2006. Full ophthalmologic examinations were conducted. Systemic clinical examinations were performed routinely with preoperative and postoperative liver function tests, chest X-ray and abdominal ultrasonography. Computed tomography or magnetic resonance imaging was used to confirm metastases that were suspected in screening examinations. All patients were evaluated at the ocular oncology clinic of Qilu hospital or Beijing Tongren hospital. The enucleation was the standard treatment for UM in China during the indicated period.
The duration of the follow-up period was calculated from the date of UM diagnosis to the date of death or last follow-up. For the purpose of the current study, patients were followed up until death or January 1st 2012, whichever came first. Eighty-nine patients with complete follow-up data have been examined in survival analyses. This study conformed to the Declaration of Helsinki and was approved by the ethics committee of the appropriate institutes. All patients had previously given their informed consent for experimental research on residual tumor tissues available after histopathologic and cytogenetic analyses.
Histopathologic examination
Histological details were obtained from the pathology reports and were reviewed and confirmed by one pathologist unaware of other data. The diagnosis of melanoma was confirmed using sections stained for hematoxylin and eosin (H&E) and/or Melan A [33]. The tumors were histologically examined for cell type, localization, size, mitosis frequency, necrosis, and scleral invasion. Spindle and epithelioid cell types were assessed using the modified Callender system [34]. Extravascular matrix patterns were assessed using the periodic acid-Schiff reagent without hematoxylin counterstaining, and the sections were viewed under a green filter [35]. The mitotic count was measured by counting the number of mitoses in 40 high-power fields (HPF) in the H&E sections [33].
Immunohistochemistry
Histological sections of formalin-fixed paraffin-embedded samples were analyzed for the presence of Ki67 and HMGA1 by the labeled streptavidin-biotin method. After deparaffinization and antigen retrieval using an autoclave oven technique, sections were incubated at 4°C overnight and incubated with rabbit polyclonal anti-HMGA1 antibody (1∶50; ab4078, Abcam Inc, Cambridge, MA) and Ki67 antigen mouse monoclonal antibody (1∶75, DakoCytomatin, Glostrup, Denmark) at 4°C. Antigen-antibody complexes were detected by the cobalt-3, 3′-diaminobenzidine reaction. Squamous cell carcinomas known to be positive for HMGA1 expression were used as positive controls [36]. Sections incubated in phosphate-buffered saline without the primary antibody served as negative controls.
Staining for HMGA1and Ki67 was considered positive when it was nuclear. Images of several high-power fields (HPF; ×400) were captured from regions with different staining intensities, including high, moderate, low, and negative staining for each case. The photographs were printed on plain paper, and a grid was drawn over them. A total of 1000 cells were counted and expressed as a percentage of tumor cells with positive nuclei. The percentage of HMGA1 positive tumor cells was scored on a scale from 0 to 4 (0, no staining; 1+, ≤10%; 2+, ≤30%; 3+, ≤50%; 4+, >50%). Then, the expression levels of HMGA1 were divided into two groups according to score: low (score: 0, 1+); high (score: 2+, 3+, and 4+) [36], [37]. The Ki67 labeling index (LI) was determined by counting the number of positive cells in a total of 800–1000 tumor cells observed in regions of highest staining (hot spot) at several HPF (×400). The results were expressed as a percentage of tumor cells with positive nuclei.
Statistical analysis
Biostatistical analyses were performed with StatView 5.0.1 software (Abacus Concepts, Berkeley, CA). The Pearson χ2 test or Fisher's exact test was used to compare qualitative variables. P values were calculated by ANOVA for age. In order to determine the significance of the associations and differences among the different variables, the data were analyzed using the Mann-Whitney U test and χ2 test. The association among the expression levels of HMGA1 and Ki67 LI were analyzed by Pearson's correlation coefficient. The Kaplan-Meier method and log-rank test were used for survival analyses. Cox proportional hazards regression model was used to compute mortality hazard ratios and 95% confidence intervals (CIs). To control for confounding variables, we used multivariate Cox proportional hazards regression models. To assess independent association between HMGA1 expression and key severity markers (epithelioid cells, mitosis count and Ki67 LI), multivariate logistic regression analysis was done and odds ratio (OR) was adjusted for age and gender. Probability values (P) <0.05 were considered to be statistically significant.
Results
Clinical and histopathologic characterisation
The 89 patients, of which 49 (55%) were male and 40 female, had a mean age of 46±14.5 years. Their tumors had a mean largest basal diameter of 14±3.4 mm (range: 7.0–21.1 mm) and a mean height of 10.1±3.3 mm (range: 3.0–20 mm). Nodular growth pattern was observed in 42 (47%) cases. There were 17 (19%) cases with ciliary body involvement, 9 (10%) cases with optic disc involvement, and 29 (32%) cases with closed vascular loops. Extraocular spread was present in 11 (12%) cases and epithelioid cell pattern was detected in 13 (15%) cases. High mitoses rates were found in 20 (24%) cases and high Ki67 labeling indexes (LIs) were detected in 19 (21%) cases (Table 1).
Table 1. Clinical, pathologic characteristics according to HMGA1 alterations in uveal melanoma.
| HMGA1 nuclear expression | ||||
| Clinical, pathologic features | Total N | High | Low | p value |
| 89 | 25 (28%) | 64 (72%) | ||
| Gender | 0.55 | |||
| Male, n (%) | 49 (55%) | 15 (60%) | 34 (51%) | |
| Female, n (%) | 40 (45%) | 10 (40%) | 30 (49%) | |
| Mean age at diagnosis ± SD | 46.0±14.5 | 48.3±15.3 | 45.1±14.2 | 0.38 |
| Laterality | 0.52 | |||
| Left eye, n (%) | 38 (43%) | 12 (48%) | 26 (41%) | |
| Right eye, n (%) | 51 (57%) | 13 (52%) | 38 (59%) | |
| Largest basal tumor diameter (mm) | ||||
| Mean (range) | 14.0 (7–21) | 14 (7–21) | 13.9 (9–18) | 0.98 |
| <15, n (%) | 59 (66%) | 17 (68%) | 42 (66%) | 0.83 |
| >15, n (%) | 30 (34%) | 8 (32%) | 22 (34%) | |
| Tumor thickness (mm) | ||||
| Mean (range) | 10.1 (3–20) | 10.2 (3–20) | 9.8 (6–15) | 0.91 |
| <10, n (%) | 46 (52%) | 14 (56%) | 32 (50%) | 0.61 |
| >10, n (%) | 43 (48%) | 11 (34%) | 32 (50%) | |
| Tumor growth pattern (Nodular) | 0.39 | |||
| Yes, n (%) | 42 (47%) | 10 (40%) | 32 (50%) | |
| No, n (%) | 47 (53%) | 15 (60%) | 32 (50%) | |
| Ciliary body involvement | 0.64 | |||
| Yes, n (%) | 17 (19%) | 4 (16%) | 13 (20%) | |
| No, n (%) | 72 (81%) | 21 (84%) | 51 (80%) | |
| Optic disc involvement | 0.71 | |||
| Yes, n (%) | 9 (10%) | 3 (13%) | 6 (9%) | |
| No, n (%) | 80 (90%) | 22 (87%) | 58 (91%) | |
| Closed loop | 0.94 | |||
| Yes, n (%) | 29 (33%) | 8 (32%) | 21 (33%) | |
| No, n (%) | 60 (67%) | 17 (68%) | 43 (67%) | |
| Extraocular spread | 0.94 | |||
| Yes, n (%) | 11 (12%) | 3 (13%) | 8 (13%) | |
| No, n (%) | 78 (88%) | 22 (87%) | 56 (87%) | |
| Epithelioid cells | 0.025 | |||
| Yes, n (%) | 13 (15%) | 7 (28%) | 6 (9%) | |
| No, n (%) | 76 (85%) | 18 (72%) | 58 (91%) | |
| Mitoses count / 40 HPF | <0.0001 | |||
| ≤4, n (%) | 69 (76%) | 11 (44%) | 58 (91%) | |
| >4, n (%) | 20 (24%) | 14 (56%) | 6 (9%) | |
| Ki67 labeling index | 0.0018 | |||
| ≤2, n (%) | 70 (79%) | 14 (56%) | 56 (87%) | |
| >2, n (%) | 19 (21%) | 11 (44%) | 8 (13%) | |
The mean follow-up time in the 89 patients that were analyzed for survival was 53±22 months (median = 55 months; range: 8–120 months), and there were 25 distant metastases and 25 deaths from metastases. The median time from enucleation to the first clinical detection of distant metastases (liver and brain) was 30 months (range: 4–56 months). The median time from metastasis to death was 5 months (range: 2–9 months). In addition, there were 5 censors with follow-up data before the reference time horizon, January 2012. However, their follow-up time was longer than 8 years.
HMGA1 expression and correlation with other characteristics in UM
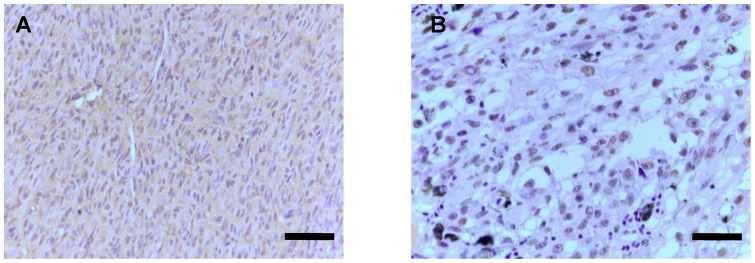
In uveal melanoma specimens, immunopositivity for HMGA1 showed a clear nuclear staining. Weak cytoplasmic localization was rarely observed. Nuclear staining was typically very intense, encompassing the entire nucleus of tumor cells, regardless of their specific type (Figure 1A, B). Nuclear expression of HMGA1 was detected in 39 uveal melanomas (44%), whereby 25 of them (28% of the total of 89 patient samples) expressed it a high levels (marked 4+, 3+ and 2+ as described in detail in Materials & Methods) (Table 1).
Figure 1. Expression of HMGA1 in UM.
Detection of high mobility group A 1 (HMGA1) immunoreactivity in UM. A, HMGA1 expression in melanoma spindle cells (Scale bar – 100µm). B, HMGA1 expression in melanoma epithelioid cells (Scale bar – 50µm).
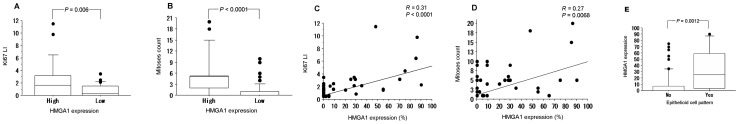
High expression levels of HMGA1 were more frequent in UM with high levels of mitoses counts and high levels of Ki67 LI (P<0.0001, P = 0.0018, respectively; Table 1). Furthermore, the Ki67 LI and mitoses counts were significantly higher in UM with high levels of HMGA1 (mean 2.4% and 5.3%, respectively) than in those with low levels of HMGA1 (median 0.7% and 1%, respectively) (high versus low, P = 0.006; and P<0.0001, respectively) (Figure 2A, B; Mann-Whitney U test). Finally, the above correlations were confirmed using Pearson's correlation coefficient analysis (R = 0.31, P<0.0001; R = 0.27, P<0.0068, respectively) (Figure 2C, D; Pearson's correlation coefficient). In addition, in a multivariate logistic regression analysis, high level of mitoses counts was independently associated with high level of HMGA1 (Table S1). The frequency of high levels HMGA1 expression was higher in UM with epithelioid cell patterns (P = 0.025; Table 1) and the mean level of HMGA1 expression (%) was also significantly higher in these cases (P = 0.0012; Figure 2E; Mann-Whitney U test). Nevertheless, there were no significant correlations between HMGA1 expression levels and other clinicopathological characteristics (Table 1).
Figure 2. Correlation of HMGA1 expression levels and other characteristics in UM.
Immunoreactivity levels of HMGA1, Ki67 LI and mitoses counts. In A, B, E, values are expressed as the median (horizontal line in each box), with the 25th and 75th percentiles (interquartile range, top and bottom of each box) and the 10th and 90th percentiles range (top and bottom of each bar). Dots indicate outliers. A, Ki67 LI was significantly higher in cases with high HMGA1 expression levels compared to cases with low expression. B, Mitosis counts were also significantly higher in HMGA1 high cases compared to HMGA1 low cases. Significant correlations have been detected between the expression of HMGA1 and Ki67 LI (C), and mitoses counts (D). E, HMGA1 expression levels were significantly higher in cases showing epithelioid cell pattern than in cases lacking this cell pattern.
Impact of HMGA1 on UM patient survival and association with distant metastases
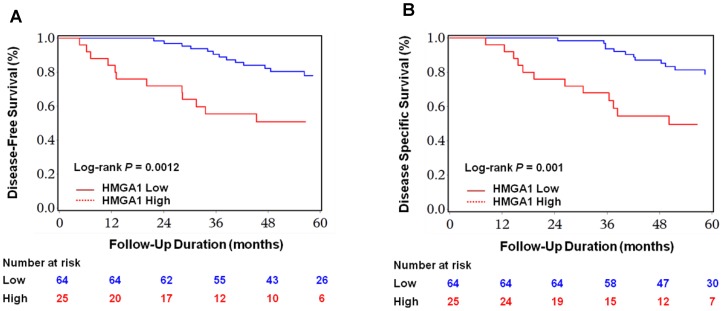
During the clinically disease-free survival period, high expression of HMGA1 was significantly correlated with increased risk of distant metastases using the log-rank test analysis (log rank P = 0.0012; Figure 3A). By univariate Cox regression, presence of epithelioid cell pattern (hazard ratio (HR), 3.02; 95% CI, 1.26–7.29; P = 0.013), high level of mitoses count (HR, 5.02; 95% CI, 2.31–9.31; P = 0.0012), high Ki67 LI (HR, 3.35; 95% CI, 1.51–6.48; P = 0.0034), and high level of HMGA1 expression (HR, 3.41; 95% CI, 1.55–7.50; P = 0.0023), were significantly associated with an increased risk of distant metastases (Table 2). By multivariate Cox regression (Table 2), both high mitoses count and high level of HMGA1 expression were significant prognostic predictors of distant metastases (multivariate HR, 4.17; 95% CI, 1.86–8.28; P = 0.005; multivariate HR, 3.44; 95% CI, 1.56–7.60; P = 0.0022; respectively).
Figure 3. Kaplan-Meier curves for survival in UM patients.
Survival of UM patients were estimated according to HMGA1 expression levels and Ki67 LI. Significant differences in disease-free survival rates were observed (A, B). Significant differences in disease-specific survival rates were also observed (C, D). P values were calculated with a log-rank test.
Table 2. Clinicopathological features, tumor markers, HMGA1, and uveal melanoma patients' survival.
| Total N (%) | No. of events | Univariate HR (95% CI) | P | Multivariate HR1 (95% CI) | P | ||
| Disease-free survival | |||||||
| Epithelioid cells | No | 76 (85%) | 18 | 1 (reference) | 1 (reference) | ||
| Yes | 13 (15%) | 7 | 3.02 (1.26 to 7.29) | 0.013 | 1.67 (0.68 to 4.25) | 0.28 | |
| Mitoses count | ≤4 | 69 (76%) | 13 | 1 (reference) | 1 (reference) | ||
| >4 | 20 (24%) | 12 | 5.02 (2.31 to 9.31) | 0.0012 | 4.17 (1.86 to 8.28) | 0.005 | |
| Ki67 LI | ≤2 | 70 (79%) | 15 | 1 (reference) | 1 (reference) | ||
| >2 | 19 (21%) | 10 | 3.35 (1.51 to 6.48) | 0.0034 | 2.12 (0.82 to 4.86) | 0.13 | |
| HMGA1 | Low | 64 (72%) | 13 | 1 (reference) | 1 (reference) | ||
| High | 25 (28%) | 12 | 3.41 (1.55 to 7.50) | 0.0023 | 3.44 (1.56 to 7.60) | 0.0022 | |
| Disease specific survival | |||||||
| Epithelioid cells | No | 76 (85%) | 18 | 1 (reference) | 1 (reference) | ||
| Yes | 13 (15%) | 7 | 3.26 (1.27 to 7.32) | 0.014 | 1.59 (0.61 to 4.12) | 0.34 | |
| Mitoses count | ≤4 | 18 (24%) | 13 | 1 (reference) | 1 (reference) | ||
| >4 | 57 (76%) | 12 | 5.12 (3.39 to 10.02) | 0.0014 | 4.15 (2.91 to 8.37) | 0.006 | |
| Ki67 LI | ≤2 | 70 (79%) | 15 | 1 (reference) | 1 (reference) | ||
| >2 | 19 (21%) | 10 | 3.52 (1.56 to 7.81) | 0.0025 | 1.99 (0.81 to 4.95) | 0.15 | |
| HMGA1 | Low | 64 (72%) | 13 | 1 (reference) | 1 (reference) | ||
| High | 25 (28%) | 12 | 3.46 (1.67 to 8.16) | 0.0013 | 2.41 (1.10 to 5.53) | 0.041 | |
Abbreviations: HR, hazard ratio; CI, confidence interval.
The multivariate Cox regression model initially included the HMGA1 expression variable (high or low), age of diagnosis, sex, largest basal tumor diameter, tumor thickness and epithelioid cell pattern.
Association with melanoma-specific survival
For disease-specific survival, high level of HMGA1 was significantly associated with an increased risk of disease specific mortality using the log-rank test analyses (P = 0.001; Figure 3B). By univariate Cox regression, presence of epithelioid cell pattern (HR, 3.26; 95% CI, 1.27–7.32; P = 0.014), high level of mitoses count (HR, 5.12; 95% CI, 3.39–10.02; P = 0.0014), high Ki67 LI (HR, 3.52; 95% CI, 1.56–7.81; P = 0.0025), and high level of HMGA1 expression (HR, 3.46; 95% CI, 1.67–8.16; P = 0.0013), were significantly associated with an increased risk of disease-specific mortality (Table 2). By multivariate Cox regression (Table 2), both high mitoses count and high level of HMGA1 expression were significant prognostic predictors of worse outcome (multivariate HR, 4.15; 95% CI, 2.91–8.37; P = 0.006; multivariate HR, 2.41; 95% CI, 1.10–5.53; P = 0.041; respectively).
Discussion
To our knowledge, this is the first study reporting HMGA1 expression in UM. In fact, nuclear expression of HMGA1 was frequent in UM, and high expression levels of HMGA1 were more prevalent in UM with high levels of epithelioid cell pattern, mitoses counts, and Ki67 LI. The latter is a cell-proliferation marker that is considered an independent prognostic parameter for UM [38]. As originally speculated high expression levels of HMGA1 protein were associated with highly malignant phenotypes of UM and hence can be considered as a marker for poor prognosis.
As metastatic UM is typically resistant to therapy [39], [40], it is critical to find methods that allow early detection of primary melanoma and that may serve to reliably predict the course of the disease. Genomic changes [16]–[19], including monosomy 3, BAP1 and somatic mutations in GNA11, are associated with metastic UM. Regarding harvesting tissue for genetic evaluation, melanoma specimens are acquired using fine needle aspiration biopsy [41]. However, currently, the two most commonly used treatments of uveal melanoma are radiation therapy and enucleation [42]. Enucleation remains the standard method of management of most large melanomas of the choroid and ciliary body. After enucleation, immunohistochemical assessment is relatively inexpensive and applicable to paraffin sections. High levels of HMGA1 expression may be linked to high proliferation rates of tumor cells by modulating chromatin structure and gene expression through DNA binding [20], [21]. There is increasing evidence suggesting that in some tumors, nuclear expression of HMGA1 was revealed a strong correlation with tumor grade and inversely associated with survival [43]–[45], HMGA1 protein can be used as a marker for metastatic progression [29]. Our previous study [28] found that high HMGA1 protein levels correlated with poor retinoblastoma prognosis. Here, we explored the correlation of HMGA1 levels with survival of UM patients. Adjusted for the largest basal tumor diameter, tumor thickness and epithelioid cell pattern, high level expression of HMGA1 was found to be associated independently with an increased risk of distant metastasis, and with shorter UM specific survival. Our data imply that HMGA1 protein is a reliable predictor for metastatic progression of UM. Furthermore, other tumor markers, such as Ki67 LI, mitoses counts and epithelioid cell pattern, which we found to be correlated with high HMGA1 expression levels in this study cohort, were also strongly correlated with metastatic mortality. This observation strengthened the role of HMGA1 protein as marker for UM metastatic prognosis.
Traditionally, prognostic markers for metastatic death from UM included the age of the patient and a largest basal diameter and an increasingly complex vascular pattern of the primary tumor [11], [14], [35], [46], [47]. A previous study [13] showed UM-specific mortality of 28.9% during a median follow-up period of 4.9 years. In our patient cohort, the mortality rate (27.8%; 24 death) during a median follow-up period of 55 months was similar despite the fact that the tumors were large (average basal diameter 14±3.4 mm) and thick (average 10.1±3.3 mm), the patients were on average younger (46±14.5 years). Evidently, the parameters used so far to predict disease outcome in this Chinese UM cohort may not be the most reliable. It is in this context that we believe that HMGA1 expression levels as determined in the present study may serve as a valuable prognostic marker.
Although much progress has been made in the diagnosis and early local treatment of UM, survival rates have not improved. Once metastases occur, prognosis is poor. In our study, as in previous ones [7], [8], the average life expectancy from diagnosis of metastasis to death was 5 months. Because HMGA1 may not only serve as a prognostic marker but may also be causally involved in metastatic progression, and because HMGA1 is not present in most adult normal tissues [20], [21], it may become an attractive target for cancer therapy [25], [48]. In fact, in pancreatic adenocarcinoma, blocking HMGA protein synthesis reduced tumor cell proliferation and metastatic potential, brought about a reduction in the Ki67 LI and caused an increase in the level of apoptosis [49]. Recently, Palmieri et al [50] reported that downregulation of HMGA-targeting miRNAs could contribute to increase HMGA protein levels in human pituitary adenomas and lead to pituitary tumorigenesis. In our previous study [28], we identified miRNAs that were dysregulated by HMGA1 in retinoblastoma, further supporting the notion that HMGA1 is not only a marker for aggressive UM but also actively involved in gene regulation in this tumor. Based on our current findings, inhibiting HMGA1 protein expression may thus offer the possibility to prevent or delay metastases of UM without damaging normal tissues.
In conclusion, high levels of HMGA1 protein correlate with poor survival and might therefore serve as a prognostic predictor of UM progression. Additional studies will show whether HMGA1 protein might provide a potential therapeutic target for preventing or delaying metastases of UM.
Supporting Information
Multivariate logistic regression analysis to assess relationship between expressions of epithelioid cells, mitoses count, Ki67 labeling index and HMGA1 status in uveal melanomas. Multivariate logistic regression analysis assessed the relationship of the presence of epithelioid cells, mitoses count, and Ki67 labeling index with HMGA1 expression status in uveal melanomas, initially included age of diagnosis and sex. * Tumor showed HMGA1-high. 1, This cut-off is same as in Table 1 and 2; 2, The median of percentage of HMGA1 expression is “0”. This cut-off is HMGA1 stained vs. non-stained; 3, 75% percentage of HMGA1 expression is “20%”. This cut-off is >20% vs. ≤20%. CI, confidence interval; OR, odds ratio.
(DOC)
Acknowledgments
The authors thank the patients who participated in the study.
Funding Statement
This research was supported in part by Shandong Science and Technology Developing Project, China (2012GSF12121), a Free Exploration Grant of Shandong University, China (2012ZD025), and Shandong Provincial Natural Science Foundation (Y2008C78). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Diener-West M, Hawkins BS, Markowitz JA, Schachat AP (1992) A review of mortality from choroidal melanoma. II. A meta-analysis of 5-year mortality rates following enucleation, 1966 through 1988. Arch Ophthalmol 110: 245–250. [DOI] [PubMed] [Google Scholar]
- 2. Egan KM, Seddon JM, Glynn RJ, Gragoudas ES, Albert DM (1988) Epidemiologic aspects of uveal melanoma. Surv Ophthalmol 32: 239–251. [DOI] [PubMed] [Google Scholar]
- 3. Singh AD, Topham A (2003) Survival rates with uveal melanoma in the United States: 1973–1997. Ophthalmology 110: 962–965. [DOI] [PubMed] [Google Scholar]
- 4. Johansson CC, Mougiakakos D, Trocme E, All-Ericsson C, Economou MA, et al. (2010) Expression and prognostic significance of iNOS in uveal melanoma. Int J Cancer 126: 2682–2689. [DOI] [PubMed] [Google Scholar]
- 5. Kujala E, Makitie T, Kivela T (2003) Very long-term prognosis of patients with malignant uveal melanoma. Invest Ophthalmol Vis Sci 44: 4651–4659. [DOI] [PubMed] [Google Scholar]
- 6. Rietschel P, Panageas KS, Hanlon C, Patel A, Abramson DH, et al. (2005) Variates of survival in metastatic uveal melanoma. J Clin Oncol 23: 8076–8080. [DOI] [PubMed] [Google Scholar]
- 7. Pyrhonen S (1998) The treatment of metastatic uveal melanoma. Eur J Cancer 34 Suppl 3S27–30. [DOI] [PubMed] [Google Scholar]
- 8. Triozzi PL, Eng C, Singh AD (2008) Targeted therapy for uveal melanoma. Cancer Treat Rev 34: 247–258. [DOI] [PubMed] [Google Scholar]
- 9. Eskelin S, Pyrhonen S, Summanen P, Hahka-Kemppinen M, Kivela T (2000) Tumor doubling times in metastatic malignant melanoma of the uvea: tumor progression before and after treatment. Ophthalmology 107: 1443–1449. [DOI] [PubMed] [Google Scholar]
- 10. Sebastian Eskelin TK (2001) Author's reply. Ophthalmology 108: 830–831. [Google Scholar]
- 11. Group COMS (2001) Assessment of metastatic disease status at death in 435 patients with large choroidal melanoma in the Collaborative Ocular Melanoma Study (COMS): COMS report no. 15. Arch Ophthalmol 119: 670–676. [DOI] [PubMed] [Google Scholar]
- 12. Coupland SE, Campbell I, Damato B (2008) Routes of extraocular extension of uveal melanoma: risk factors and influence on survival probability. Ophthalmology 115: 1778–1785. [DOI] [PubMed] [Google Scholar]
- 13.Damato B, Coupland SE (2009) A reappraisal of the significance of largest basal diameter of posterior uveal melanoma. Eye (Lond) 23: 2152–2160; quiz 2161–2152. [DOI] [PubMed]
- 14. Folberg R, Pe'er J, Gruman LM, Woolson RF, Jeng G, et al. (1992) The morphologic characteristics of tumor blood vessels as a marker of tumor progression in primary human uveal melanoma: a matched case-control study. Hum Pathol 23: 1298–1305. [DOI] [PubMed] [Google Scholar]
- 15. McLean IW, Zimmerman LE, Evans RM (1978) Reappraisal of Callender's spindle a type of malignant melanoma of choroid and ciliary body. Am J Ophthalmol 86: 557–564. [DOI] [PubMed] [Google Scholar]
- 16. Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, et al. (2010) Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 330: 1410–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prescher G, Bornfeld N, Hirche H, Horsthemke B, Jockel KH, et al. (1996) Prognostic implications of monosomy 3 in uveal melanoma. Lancet 347: 1222–1225. [DOI] [PubMed] [Google Scholar]
- 18. Trolet J, Hupe P, Huon I, Lebigot I, Decraene C, et al. (2009) Genomic profiling and identification of high-risk uveal melanoma by array CGH analysis of primary tumors and liver metastases. Invest Ophthalmol Vis Sci 50: 2572–2580. [DOI] [PubMed] [Google Scholar]
- 19. Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, et al. (2010) Mutations in GNA11 in uveal melanoma. N Engl J Med 363: 2191–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chiappetta G, Avantaggiato V, Visconti R, Fedele M, Battista S, et al. (1996) High level expression of the HMGI (Y) gene during embryonic development. Oncogene 13: 2439–2446. [PubMed] [Google Scholar]
- 21. Zhou X, Benson KF, Ashar HR, Chada K (1995) Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 376: 771–774. [DOI] [PubMed] [Google Scholar]
- 22. Wood LJ, Maher JF, Bunton TE, Resar LM (2000) The oncogenic properties of the HMG-I gene family. Cancer Res 60: 4256–4261. [PubMed] [Google Scholar]
- 23. Wood LJ, Mukherjee M, Dolde CE, Xu Y, Maher JF, et al. (2000) HMG-I/Y, a new c-Myc target gene and potential oncogene. Mol Cell Biol 20: 5490–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reeves R, Edberg DD, Li Y (2001) Architectural transcription factor HMGI(Y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol Cell Biol 21: 575–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fedele M, Pentimalli F, Baldassarre G, Battista S, Klein-Szanto AJ, et al. (2005) Transgenic mice overexpressing the wild-type form of the HMGA1 gene develop mixed growth hormone/prolactin cell pituitary adenomas and natural killer cell lymphomas. Oncogene 24: 3427–3435. [DOI] [PubMed] [Google Scholar]
- 26. Xu Y, Sumter TF, Bhattacharya R, Tesfaye A, Fuchs EJ, et al. (2004) The HMG-I oncogene causes highly penetrant, aggressive lymphoid malignancy in transgenic mice and is overexpressed in human leukemia. Cancer Res 64: 3371–3375. [DOI] [PubMed] [Google Scholar]
- 27. Fusco A, Fedele M (2007) Roles of HMGA proteins in cancer. Nat Rev Cancer 7: 899–910. [DOI] [PubMed] [Google Scholar]
- 28. Mu G, Liu H, Zhou F, Xu X, Jiang H, et al. (2010) Correlation of overexpression of HMGA1 and HMGA2 with poor tumor differentiation, invasion, and proliferation associated with let-7 down-regulation in retinoblastomas. Hum Pathol 41: 493–502. [DOI] [PubMed] [Google Scholar]
- 29. Shah SN, Resar LM (2012) High mobility group A1 and cancer: potential biomarker and therapeutic target. Histol Histopathol 27: 567–579. [DOI] [PubMed] [Google Scholar]
- 30. Takaha N, Sowa Y, Takeuchi I, Hongo F, Kawauchi A, et al. (2012) Expression and role of HMGA1 in renal cell carcinoma. J Urol 187: 2215–2222. [DOI] [PubMed] [Google Scholar]
- 31. Chuma M, Saeki N, Yamamoto Y, Ohta T, Asaka M, et al. (2004) Expression profiling in hepatocellular carcinoma with intrahepatic metastasis: identification of high-mobility group I(Y) protein as a molecular marker of hepatocellular carcinoma metastasis. Keio J Med 53: 90–97. [DOI] [PubMed] [Google Scholar]
- 32. Scholzen T, Gerdes J (2000) The Ki-67 protein: from the known and the unknown. J Cell Physiol 182: 311–322. [DOI] [PubMed] [Google Scholar]
- 33. Damato B, Duke C, Coupland SE, Hiscott P, Smith PA, et al. (2007) Cytogenetics of uveal melanoma: a 7-year clinical experience. Ophthalmology 114: 1925–1931. [DOI] [PubMed] [Google Scholar]
- 34. McLean IW, Foster WD, Zimmerman LE, Gamel JW (1983) Modifications of Callender's classification of uveal melanoma at the Armed Forces Institute of Pathology. Am J Ophthalmol 96: 502–509. [DOI] [PubMed] [Google Scholar]
- 35. Folberg R, Rummelt V, Parys-Van Ginderdeuren R, Hwang T, Woolson RF, et al. (1993) The prognostic value of tumor blood vessel morphology in primary uveal melanoma. Ophthalmology 100: 1389–1398. [DOI] [PubMed] [Google Scholar]
- 36. Wang EL, Qian ZR, Rahman MM, Yoshimoto K, Yamada S, et al. (2010) Increased expression of HMGA1 correlates with tumour invasiveness and proliferation in human pituitary adenomas. Histopathology 56: 501–509. [DOI] [PubMed] [Google Scholar]
- 37. Rahman MM, Qian ZR, Wang EL, Sultana R, Kudo E, et al. (2009) Frequent overexpression of HMGA1 and 2 in gastroenteropancreatic neuroendocrine tumours and its relationship to let-7 downregulation. Br J Cancer 100: 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mooy CM, de Jong PT, Van der Kwast TH, Mulder PG, Jager MJ, et al. (1990) Ki-67 immunostaining in uveal melanoma. The effect of pre-enucleation radiotherapy. Ophthalmology 97: 1275–1280. [DOI] [PubMed] [Google Scholar]
- 39. Augsburger JJ, Correa ZM, Shaikh AH (2009) Effectiveness of treatments for metastatic uveal melanoma. Am J Ophthalmol 148: 119–127. [DOI] [PubMed] [Google Scholar]
- 40. Kim IK, Lane AM, Gragoudas ES (2010) Survival in patients with presymptomatic diagnosis of metastatic uveal melanoma. Arch Ophthalmol 128: 871–875. [DOI] [PubMed] [Google Scholar]
- 41. Shields CL, Ganguly A, Bianciotto CG, Turaka K, Tavallali A, et al. (2011) Prognosis of uveal melanoma in 500 cases using genetic testing of fine-needle aspiration biopsy specimens. Ophthalmology 118: 396–401. [DOI] [PubMed] [Google Scholar]
- 42. Griewank KG, Murali R (2013) Pathology and genetics of uveal melanoma. Pathology 45: 18–27. [DOI] [PubMed] [Google Scholar]
- 43. Flohr AM, Rogalla P, Bonk U, Puettmann B, Buerger H, et al. (2003) High mobility group protein HMGA1 expression in breast cancer reveals a positive correlation with tumour grade. Histol Histopathol 18: 999–1004. [DOI] [PubMed] [Google Scholar]
- 44. Hristov AC, Cope L, Di Cello F, Reyes MD, Singh M, et al. HMGA1 correlates with advanced tumor grade and decreased survival in pancreatic ductal adenocarcinoma. Mod Pathol 23: 98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sarhadi VK, Wikman H, Salmenkivi K, Kuosma E, Sioris T, et al. (2006) Increased expression of high mobility group A proteins in lung cancer. J Pathol 209: 206–212. [DOI] [PubMed] [Google Scholar]
- 46. Diener-West M, Reynolds SM, Agugliaro DJ, Caldwell R, Cumming K, et al. (2004) Screening for metastasis from choroidal melanoma: the Collaborative Ocular Melanoma Study Group Report 23. J Clin Oncol 22: 2438–2444. [DOI] [PubMed] [Google Scholar]
- 47. Gragoudas E, Li W, Goitein M, Lane AM, Munzenrider JE, et al. (2002) Evidence-based estimates of outcome in patients irradiated for intraocular melanoma. Arch Ophthalmol 120: 1665–1671. [DOI] [PubMed] [Google Scholar]
- 48. Scala S, Portella G, Fedele M, Chiappetta G, Fusco A (2000) Adenovirus-mediated suppression of HMGI(Y) protein synthesis as potential therapy of human malignant neoplasias. Proc Natl Acad Sci U S A 97: 4256–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liau SS, Jazag A, Whang EE (2006) HMGA1 is a determinant of cellular invasiveness and in vivo metastatic potential in pancreatic adenocarcinoma. Cancer Res 66: 11613–11622. [DOI] [PubMed] [Google Scholar]
- 50. Palmieri D, D'Angelo D, Valentino T, De Martino I, Ferraro A, et al. (2012) Downregulation of HMGA-targeting microRNAs has a critical role in human pituitary tumorigenesis. Oncogene 31: 3857–3865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multivariate logistic regression analysis to assess relationship between expressions of epithelioid cells, mitoses count, Ki67 labeling index and HMGA1 status in uveal melanomas. Multivariate logistic regression analysis assessed the relationship of the presence of epithelioid cells, mitoses count, and Ki67 labeling index with HMGA1 expression status in uveal melanomas, initially included age of diagnosis and sex. * Tumor showed HMGA1-high. 1, This cut-off is same as in Table 1 and 2; 2, The median of percentage of HMGA1 expression is “0”. This cut-off is HMGA1 stained vs. non-stained; 3, 75% percentage of HMGA1 expression is “20%”. This cut-off is >20% vs. ≤20%. CI, confidence interval; OR, odds ratio.
(DOC)