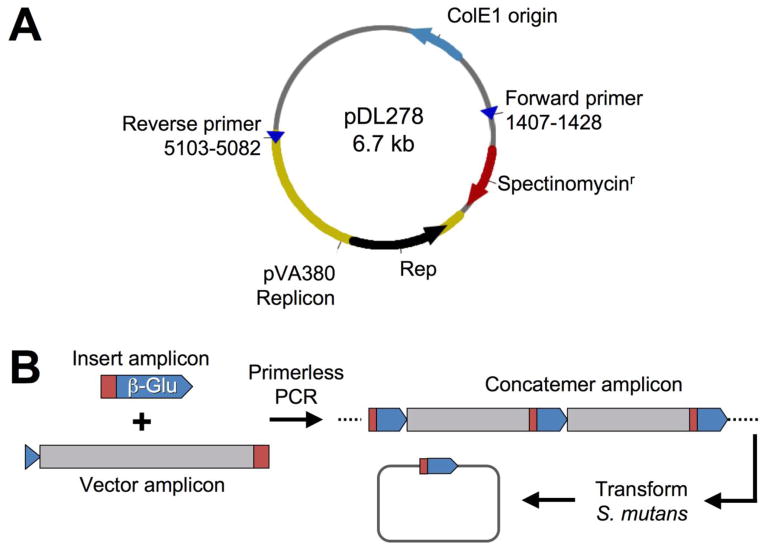
Fig. 1. Protocol for creating replicating plasmids in S. mutans using POE-PCR.
(A) The E. coli – Streptococcus shuttle vector pDL278 is shown. The indicated primers were used to PCR amplify the streptococcal origin of replication together with the spectinomycin resistance cassette. (B) Illustration of the POE-PCR procedure used to create β-glucuronidase expressing plasmids. The target vector was amplified from the shuttle vector pDL278, while the transcription fusion to the gusA gene was PCR amplified from an existing S. mutans gusA reporter strain. The primers used to amplify the gusA transcription fusion also contained complementary regions to the intended insertion site of the target vector (represented in red and blue). The two resulting PCR amplicons were subsequently mixed in a similar molar ratio and then assembled into linear concatemers through a second round of PCR. The regions of complementarity between the insert and vector amplicons served as the primers in the final PCR. Thus, no additional primers were added to this reaction. The resulting mixture of PCR generated concatemers was transformed directly into S. mutans, where the concatemers were recircularized into replicating plasmids containing the desired insert.