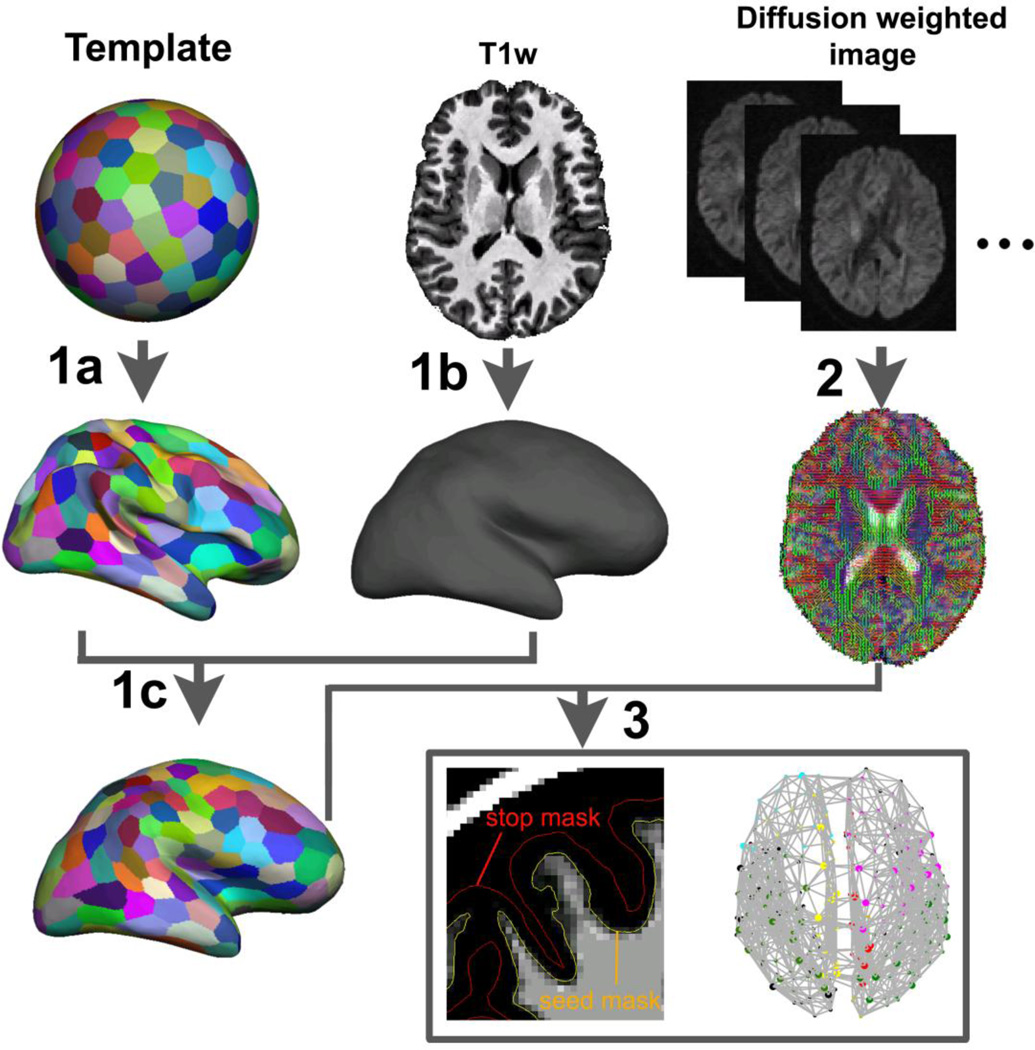
Fig. 2.
Flowchart of deriving whole-brain anatomical brain networks. The procedures could be divided into three steps: First, a surface template was generated on a sphere space and randomly parcellated into 300 and 600 parcels with the medial wall excluded. The parcellation schemes were transformed to the template original surface (1a). Each subject’s original WM/GM boundary surface was constructed based on the T1-weighted images using the FreeSurfer (1b). Then the parcellation schemes were transformed to each subject’s surface space (1c). In the second step, local diffusion orientation distribution functions in each voxel of subject’s diffusion data were estimated using FDT toolbox (2). Then the connections between each cortical parcel pairs were reconstructed by sending millions of samples from vertices on the WM/GM interface surface and the number of the “probabilistic streamlines” sent from every vertex and reached every other vertices were counted and summed over parcels for graph-theoretic analyses (3).