Abstract
Background
Hypertension represents a complex heritable disease in which environmental factors may directly affect gene function via epigenetic mechanisms. The aim of this study was to test the hypothesis that dietary salt influences the activity of a histone modifying enzyme, lysine-specific demethylase 1 (LSD-1), which in turn is associated with salt-sensitivity of blood pressure (BP).
Methods
Animal and human studies were performed. Salt-sensitivity of LSD-1 expression was assessed in wild-type and LSD-1 heterozygote knockout (LSD-1+/−) mice. Clinical relevance was tested by multivariate associations between single nuclear polymorphisms (SNPs) in the LSD-1 gene and salt-sensitivity of BP, with control of dietary sodium, in a primary African-American hypertensive cohort and two replication hypertensive cohorts (Caucasian and Mexican-American).
Results
LSD1 expression was modified by dietary salt in wild-type mice with lower levels associated with liberal salt intake. LSD-1+/− mice expressed lower LSD-1 protein levels than wild-type mice in kidney tissue. Similar to LSD-1+/− mice, African-American minor allele carriers of two LSD-1 SNPs displayed greater change in systolic BP in response to change from low to liberal salt diet (rs671357, p=0.01; rs587168, p=0.005). This association was replicated in the Hispanic (rs587168, p=0.04) but not the Caucasian cohort. Exploratory analyses demonstrated decreased serum aldosterone concentrations in African-American minor allele carriers similar to findings in the LSD-1+/− mice, decreased alpha-EnaC expression in LSD-1+/− mice, and impaired renovascular responsiveness to salt loading in minor allele carriers.
Conclusion
The results of this translational research study support a role for LSD1 in the pathogenesis of salt-sensitive hypertension.
Keywords: Hypertension, Salt-sensitivity, LSD1, Genetics, Epigenetic
INTRODUCTION
Essential hypertension (HTN) is a complex heritable disease. The ability to translate gene-based knowledge into improved treatment strategies has been disappointing. In addition to inherent heterogeneity, lack of success may be due to a failure to recognize important effects of environmental factors on gene expression (i.e. epigenetics). Dietary salt 1–3 is one such factor, yet little is known of its effect on gene expression in blood pressure (BP) regulation. This is of particular importance in African-American hypertensives who have a high prevalence of salt-sensitivity and cardiovascular risk.
In an animal model, we recently identified that lysine-specific demethylase 1 (LSD-1) is involved in salt-sensitive HTN4. LSD-1 acts as a regulator of gene transcription through alteration in histone methylation5–10. The heterozygote knockout of LSD-1 (LSD-1+/−) demonstrates, 1) decreased LSD-1 mRNA and protein levels in heart and aorta, 2) salt-sensitivity of BP, 3) lower plasma and urine aldosterone levels, 4) and altered aortic vasoactivity4. This report expands on these results in four ways. First, are LSD-1 levels reduced in the kidney similar to the heart/vasculature? Second, in the setting of reduced aldosterone, what are the renal epithelial sodium channel (ENaC) levels given its role in salt handling? Third, does sodium intake change LSD-1 levels? Finally, to translate the clinical relevance of these findings, we evaluate the relationship of polymorphic variants in the LSD-1 gene to salt-sensitivity in human HTN, focusing initially on African-Americans in light of their inherent salt-sensitivity3, 11–14.
METHODS
Animal Studies
Animals
The interaction of salt intake and LSD-1 was evaluated in wild-type (WT) and LSD-1+/− mice treated with a liberal sodium (HS) (1.6% Na+) or low sodium (LS) (0.02% Na+) diet for one week. LSD-1+/− mice were generated using a gene trap approach as previously described4. The LSD-1 homozygote knockout is embryonically lethal. As previously described, the LSD-1+/− mice compared to WT, have similar systolic BP (SBP) on LS, and significantly higher SBP on HS intake (p<0.001). LSD-1+/− mice had similar total body and kidney weights but significantly heavier hearts (difference = 50 mg; p=0.02)4.
Animals were housed in an animal facility with 12 hr light/dark cycle, at an ambient temperature of 22±1°C. All studies were performed between 7–9 AM. All experimental procedures were approved by the Institutional Animal Care and Use Committee at Harvard Medical School.
Tissue preparation
Mice were euthanized under deep anesthesia with isofluorane, the abdominal cavity opened, and renal cortical tissue excised and placed in liquid nitrogen in preparation for protein analysis.
Western blot analysis
Protein was extracted by homogenizing tissue with RIPA lysis buffer (Santa Cruz Biotechnology Inc, Santa Cruz, CA) as previously described4. Protein extracts (40 μg) were combined with equal volume 2X Laemmli loading buffer, boiled for 5 min, and size-fractionated by electrophoresis on 7.5% SDS-polyacrylamide gels. Proteins were transferred from gel to a nitrocellulose membrane by electroblotting. Membranes were incubated with 5% non-fat dried milk in TBS-Tween (USB Corporation, Cleveland, OH) for 1h and incubated overnight at 4°C with primary antibodies. After incubation, samples were washed, incubated with peroxidase-conjugated secondary antibody, and analyzed using enhanced chemiluminescence (Perkin-Elmer Life Sciences, Boston, MA). Blots were subsequently re-probed for β-actin (1:5000 dilution) and results normalized to correct for loading. Immunoreactive bands were analyzed quantitatively by optical densitometry, and densitometry values represented pixel intensity. Primary antibodies used were from Abcam: rabbit anti-LSD1 (catalog number ab17721; dilution 1:1000), from Novus Biologicals : rabbit anti-ENaC (dilution 1:200); and from Sigma-Aldrich: mouse anti-beta-actin (catalog number A5441; dilution 1:5000).
Western blots included a negative control lane to verify antibodies did not bind non-specifically. Moreover, a marked decrease (>90%) in LSD-1 levels was observed by Western blot after LSD-1 had been knocked down with siRNA (not shown). No positive controls were included. LSD-1 antibody did not bind to the mineralocorticoid receptor (MR), angiotensin-II (AngII) receptor, angiotensin converting enzyme (ACE).
Human Studies
To augment the association with salt-sensitivity, we chose an African-American hypertensive population from HyperPATH (HyperPATH-AA; n=63; Table I) as our primary cohort. Replication cohorts included 422 Caucasian hypertensives from HyperPATH (HyperPATH-C) and 178 Mexican-American hypertensives from HTN-IR. Protocols for subject recruitment and data collection for HyperPATH and HTN-IR populations have been previously described15, 16. The analyses were restricted to subjects with LSD-1 genotype data. The institutional review board of each site approved the study. All subjects gave written informed consent before enrollment. Data presented have not been previously reported except as an abstract17.
Table I.
Baseline Characteristics of the Primary HyperPATH-AA Cohort
| African Americans | |||
|---|---|---|---|
| Major Allele Homozygotes | Minor Allele Carriers | ||
| Age | 45.8 ± 1.4 | 46.7 ± 1.4 | |
| Gender (% female) | 50 | 50 | |
| BMI (kg/m2) | 28.7 ± 1.0 | 28.8 ± 0.8 | |
| SBP (mmHg) | HS | 149.7 ± 4.1 | 155.4 ± 3.7 |
| LS | 138.3 ± 3.6 | 134.9 ± 2.9 | |
| DBP (mmHg) | HS | 87.3 ± 1.9 | 90.7 ± 2.1 |
| LS | 81.5 ± 2.1 | 81.97 ± 2.3 | |
| Urine Sodium (mmol/24-hr) | HS | 217.7 ± 16.9 | 212.2 ± 12.5 |
| LS | 15.7 ± 1.8 | 16.5 ± 4.1 | |
| PRA (ng/ml/hr) | HS | 0.35 ± 0.05 | 0.25 ± 0.03 |
| LS | 2.64 ± 0.90 | 1.51 ± 0.31 | |
| Serum Aldo (ng/dl) | HS | 5.94 ± 0.89 | 3.44 ± 0.32* |
| LS | 15.02 ± 1,5 | 13.66 ± 1.1 | |
| Serum AngII (pg/ml) | HS | 25.19 ± 3.7 | 18.14 ± 3.2 |
| LS | 32.73 ± 4.6 | 27.05 ± 3.3 | |
| RBF (cc/min/1.73 m2) | HS | 519.51 ± 21.3 | 574.94 ± 42.3 |
| LS | 501.00 ± 25.1 | 572.96 ± 45.6 | |
Data shown are baseline unadjusted values for SNP rs587168 on liberal sodium (HS) and low-sodium (LS) diets. Values are reported as mean ± SEM (except % for gender).
= p<0.05
BMI=body mass index; SBP=systolic blood pressure; DBP=diastolic blood pressure; PRA=plasma renin activity; Aldo=aldoseterone; AngII=angiotensin-II; RBF=renal blood flow
HyperPATH Subjects and Protocol
The HyperPATH consortium represents an intensive phenotyping program to determine the genetic underpinnings of HTN. Its strengths include an enhanced signal-to-noise ratio through rigorous control of factors that influence BP including medication washout, body positioning, diurnal variation, and dietary salt. Subjects were studied in Clinical Research Centers (CRC) of five sites (Brigham and Women’s Hospital, Boston, USA; University of Utah Medical Center, Salt Lake City, USA; Vanderbilt University, Nashville, USA; Hospital Broussais, Paris, France and University La Sapienza, Rome, Italy). The HyperPATH inclusion/exclusion criteria and the detailed phenotyping protocol have been previously described15, 18. As per the original study design, HTN was defined as seated diastolic BP ≥100 mmHg off antihypertensive medication, ≥90mmHg on at least one antihypertensive medication, or treatment with up to three antihypertensive medications. Use of more than three medications was exclusionary.
All subjects completed a screening visit consisting of a physical examination, medical history, and laboratory assessment. A history of diabetes mellitus, secondary HTN, renal insufficiency (serum creatinine >1.5mg/dL), other significant medical illnesses, current tobacco or illicit drug use, or alcohol intake >12oz per week was exclusionary. Normal laboratory values for electrolytes, thyroid and liver function tests were required. Subjects were between 18–65 years of age. Race was self-defined. To minimize interference of medication with renin-angiotensin-aldosterone system (RAAS) activity, all ACE inhibitors, angiotensin receptor blockers and MR antagonists were discontinued for 3 months. If needed, subjects were placed on amlodipine or hydrochlorothiazide to control BP. At three weeks prior to hormonal and vascular assessment all antihypertensive medications were discontinued.
All subjects completed two controlled dietary phases: five to seven days of HS (200 mmol Na+/day) and seven days of LS (10 mmol Na+/day) diet, prepared by each site’s metabolic kitchen. Each diet was isocaloric, contained 100 mmol/day potassium, 20 mmol/day calcium, and caffeine and alcohol-free. On the final day of each diet, subjects were admitted to the CRC and maintained fasting and supine overnight. Measurement of sodium and creatinine excretion in a 24-hour urine collection confirmed sodium balance (≥150 mmol for HS and <30 mmol for LS). Hemodynamic and laboratory assessments were obtained in supine posture between 8AM–10AM. BP was measured at five-minute intervals using an automated device (DINAMAP; Critikon, Tampa, FL). Three consecutive readings were averaged for analysis. Salt-sensitivity of BP was recorded as a continuous variable representing the change in SBP in response to dietary salt (ΔSBP). A subset of African-American subjects (n=22) also completed assessment of effective renal blood flow (RBF) by para-aminohippuric acid clearance method19.
HTN-IR Subjects and Protocol
The Mexican-American Hypertension cohort consists of Hispanic families (939 individuals from 160 pedigrees) ascertained via a hypertensive proband (HTN defined as sitting BP≥140/90 mm Hg off medication)16. Probands were recruited through the HTN Clinic at Los Angeles County, University of Southern California Medical Center or the CRC at the University of California, Los Angeles. Subjects were 18–65 years, with mild-moderate HTN and no target organ damage. Antihypertensive medications were discontinued for 2 weeks prior to study if permissible. Salt-sensitivity of BP protocols included the Weinberger method20, which involved 1) infusion of 1 L of 0.9% saline over 4h on day 1; and 2) 10mEq sodium diet and 3 doses of furosemide on day 2. BP was measured at the end of both procedures with the difference (salt loaded - salt depleted) representing measure of salt-sensitivity (SBPsen).
Phenotype(s) Examined
The primary phenotype was salt-sensitivity defined as ΔSBP in HyperPATH and SBPsen in HTN-IR as described above. Aldosterone levels, plasma renin activity (PRA) and RBF were examined as secondary phenotypes in an exploratory analysis of underlying mechanisms.
Laboratory Analyses
HyperPATH samples were analyzed at a central laboratory. Samples were collected on ice and centrifuged for 20 minutes. Serum and urine specimens were stored at −20°C without preservatives until assayed. Sodium and potassium levels were measured by flame photometry. Aldosterone and renin levels were measured by radioimmunoassay techniques21, 22.
Genotyping and SNP Selection
DNA was extracted from peripheral leukocytes using standard procedures. In HyperPATH, genotyping was conducted using the Illumina Bead Station Golden Gate platform. Tagging single nucleotide polymorphisms (SNPs) were identified from Haploview using the HapMap23 CEU and YRI populations with an R2 > 0.9 and a minor allele frequency (MAF) >10%. This provided 7 tagging-SNPs (Table II). All SNPs had a completion rate of >95%. Repeat genotyping for 10% of the SNPs demonstrated concordance with the original genotype call.
Table II.
Allele Frequency Data for HapMap Tagging SNPs of LSD1 in the HyperPATH African-American Cohort
| African American Cohort | ||
|---|---|---|
| SNP | MAF | HWE p-value |
| rs681648† | 0.159 | 0.64 |
| rs671357† | 0.14 | 0.42 |
| rs6683468 | 0.11 | 0.34 |
| rs7548692 | 0.33 | 0.998 |
| rs10799789* | 0.29 | 0.91 |
| rs587168* | 0.278 | 0.82 |
| rs10489564 | 0.206 | 0.09 |
MAF=minor allele frequency, HWE=Hardy Weinberg Equilibrium
and † represent SNPs in linkage disequilibrium with each other
Linkage disequilibrium (LD) constructs in our HyperPATH-AA population revealed that 2 of 7 tagging-SNPs were in LD with other genotyped SNPs, and 5 of 7 SNPs captured 100% of the common LSD1 variation with an r2 >0.85. Therefore, 5 SNPs were analyzed in the HyperPATH-AA cohort. The SNP most significantly associated with salt-sensitivity in HyperPATH-AA (rs587168) was examined in HyperPATH-C and HTN-IR. In the HTN-IR cohort, genotyping was performed using a 7600-SNP Illumina iSelect platform.
Statistical Analysis
All tests for association were performed using SAS 9.1 (SAS Institute, Cary, NC). Hardy-Weinberg Equilibrium was assessed using the chi-square test. The D′ and r2 measures of LD were estimated using Haploview. In the HyperPATH cohorts, a mixed effect linear regression model (PROC MIXED) was used to test associations between LSD-1 genotype and phenotypes, accounting for relatedness, and adjusted for age, gender, body mass index (BMI) and study site. Logarithmic transformations were performed for serum aldosterone and PRA to meet normality assumptions. Wilcoxon Signed Ranks test was used for paired comparison of RBF on HS versus LS diet within genotype. In the HTN-IR cohort, the PROC GENMOD procedure in SAS was used for analyzing genotype-phenotype association utilizing the generalized estimating equation, accounting for familial correlations and adjusted for gender, age and BMI.
For the primary phenotype (ΔSBP), p = 0.01 was considered statistically significant in the HyperPATH-AA cohort to account for testing 5 SNPs. Given that the AA population could be highly admixed, we tested for population stratification. Using the control SNPs available in this cohort (N=801), the genomic inflation factor was 1.007. Statistical significance was indicated by p <0.05 for all secondary analyses and replication analyses in the HyperPATH-C and HTN-IR cohorts, as only one SNP was tested. For animal studies, data were analyzed by Student’s t-test using Graphpad Prism, with p <0.05 considered significant.
RESULTS
Relationship between LSD-1 and Dietary Sodium
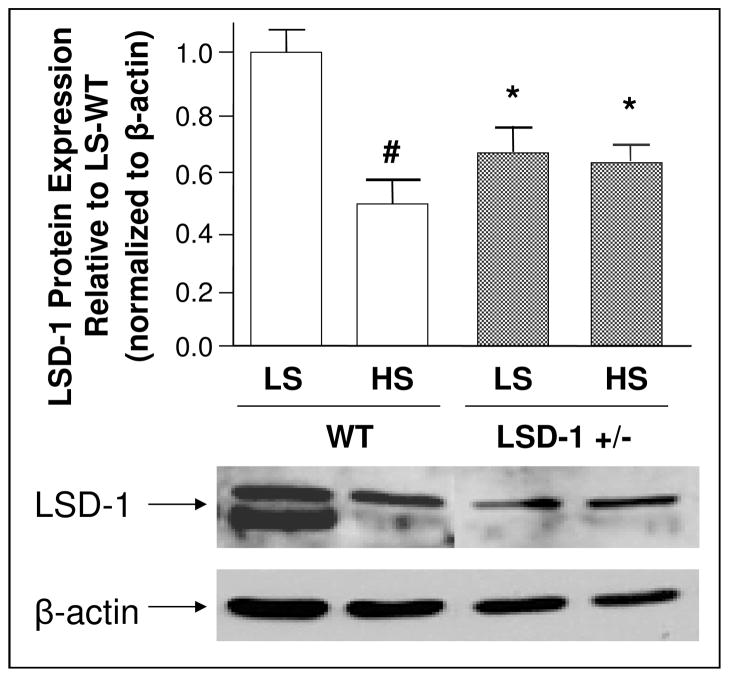
LSD-1 protein levels were significantly higher on LS compared to HS diet in WT mouse kidney tissue (Figure 1). In contrast, LSD-1 protein levels did not respond to salt modulation in LSD-1+/− mice (Figure 1). A similar change in LSD-1 protein was seen in mouse heart tissue (data not shown).
Figure 1. Effect of dietary sodium on LSD-1 protein levels in mouse kidney.
# LSD-1 protein levels were significantly higher on low salt (LS) compared to liberal salt (HS) diet in kidney tissue of wild-type (WT) mice (N=3–4; p < 0.01).
* In LSD-1 heterozygote knockout (LSD-1+/−) mice, LSD-1 protein levels in the kidney were significantly lower compared to WT mice on both diets (N=3–4; p < 0.05).
LSD-1 protein levels in the LSD-1+/− mice did not change significantly between diets. Error bars = SEM.
Baseline Characteristics of Human Subjects
Characteristics of the primary HyperPATH-AA cohort are shown in Table 1. Baseline characteristics (age, BMI, gender) did not differ greatly between study cohorts.
Relationship between Salt-Sensitivity of BP and LSD-1 Genotype in Human Hypertension
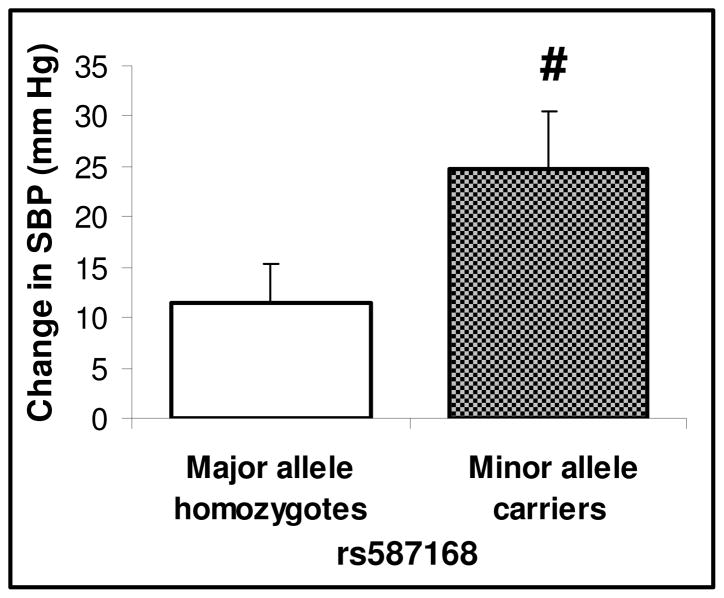
All SNPs were in Hardy-Weinberg Equilibrium. In the HyperPATH-AA cohort, a significant association was observed for ΔSBP in 2 of 5 SNPs (rs671357, p=0.01; rs587168, p<0.005), whereby minor allele carriers displayed a greater ΔSBP (Figure 2). To replicate this finding, we tested SNP rs587168 in the HyperPATH-C and HTN-IR cohorts. In the HTN-IR cohort, minor allele homozygotes of rs587168 had significantly greater SBPsen (11.11 ± 2.58 (mean±SEM) mmHg in minor allele homozygotes versus 5.49 ± 1.23 mmHg in major allele carriers, p=0.04). No association with ΔSBP was observed in Caucasians (14 ± 1 mmHg in minor allele carriers versus 15 ± 1 mmHg in major allele homozygotes, p=0.25).
Figure 2. Effect of LSD-1 genetic variants on the systolic blood pressure (SBP) response to dietary salt in African-American Hypertensive Subjects.
# African-American hypertensive minor allele carriers of LSD-1 rs587168 (N=26) compared with major allele homozygotes (N=28) had significantly greater increase in SBP in response to change from low salt to liberal salt diet (p < 0.01). Data shown are mean values adjusted for age, gender and BMI differences. Error bars = SEM.
A significant association of the minor allele with salt-sensitivity of BP was also seen for rs671357 (data not shown; p = 0.01) in the African-American cohort.
Exploratory Mechanistic Studies in Humans and Mice
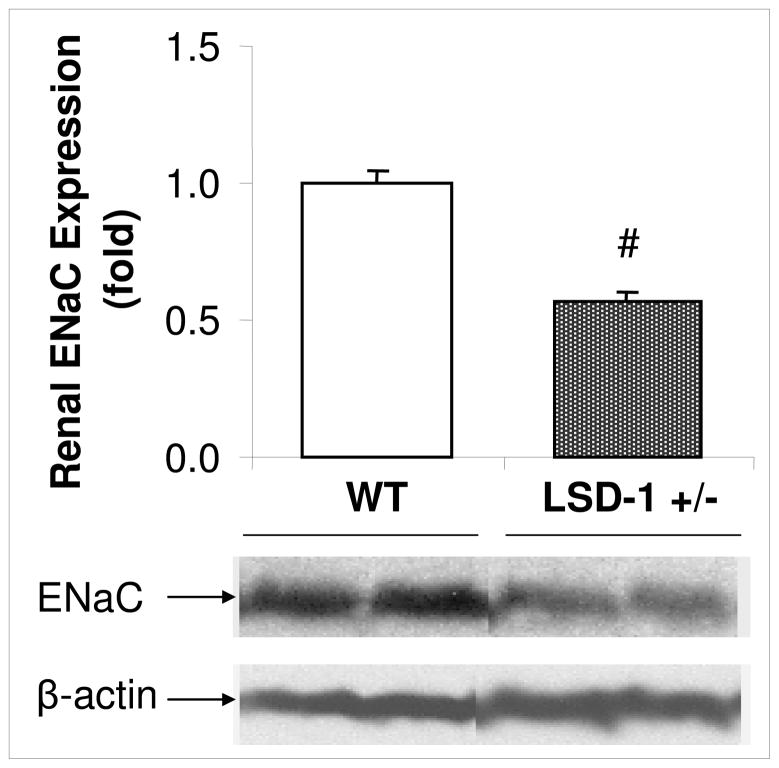
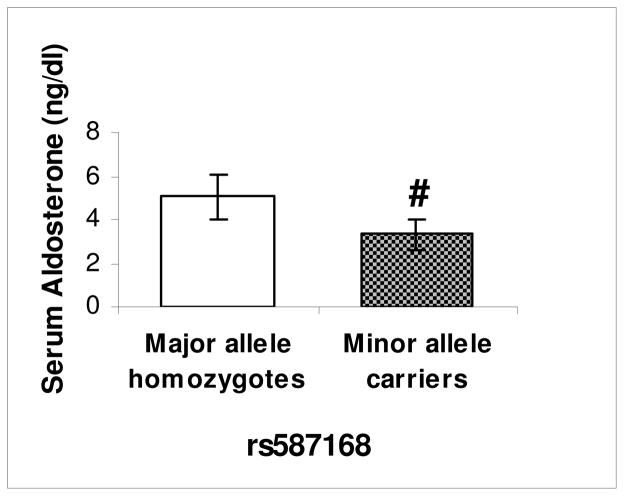
To identify potential mechanisms, we assessed physiologic pathways involved in salt homeostasis. We previously reported that LSD-1+/− have lower serum aldosterone and 24-hour aldosterone excretion compared with WT mice4. We now assessed whether a target of aldosterone linked to sodium reabsorption, renal alpha-ENaC, may be dysfunctional. Consistent with lower aldosterone, LSD-1+/− mice had lower alpha-ENaC expression compared to WT (Figure 3). Concordant with animal data, HyperPATH-AA minor allele carriers of rs587168 had lower serum aldosterone on HS diet compared to major allele homozygotes (Figure 4; p=0.003). No significant differences were observed in serum aldosterone on LS or PRA on either diet between genotype, although there was a trend towards lower PRA in the minor allele (Table I). We also assessed for correlation between SBP and PRA, AngII or aldosterone by genotype in the HyperPATH-AA cohort. As might be anticipated given the small sample size, we found none except a marginal correlation (p=0.05) between aldosterone and SBP on HS diet. We then explored the relationship between salt and RBF by LSD-1 genotype. Minor allele carriers for rs587168 (n=12) had no change in RBF (1.99 ± 22.7 (mean±SEM) cc/min/1.73 m2) in response to change from LS to HS diet whereas major allele homozygotes (n=10) had an increase in RBF (18.5 ± 24.3 cc/min/1.73 m2) as would be expected in response to increased salt intake, although not statistically significant.
Figure 3. ENaC expression on liberal salt diet.
# LSD-1 heterozygote knockout (LSD-1+/−) compared with wild-type (WT) mice, had significantly lower expression of the alpha subunit of renal epithelial sodium channel (ENaC) (N=3–4 mice in each group; p < 0.01) on liberal salt diet. Error bars=SEM.
Figure 4. Aldosterone levels by LSD1 genotype.
# African-American hypertensive minor allele carriers of rs587168 (N=26) had significantly lower serum aldosterone levels compared with major allele homozygotes (N=28) on liberal salt diet (p <0.01). Error bars = 95% confidence interval.
DISCUSSION
We report that dietary salt modifies LSD-1 expression and that genetic alteration in LSD-1 (likely “loss of function”, although a dominant negative effect can not be ruled out) is associated with salt-sensitivity of BP in both an animal model and human HTN. This finding is important because it provides evidence for an environmental factor (dietary salt) affecting expression of a gene known to be an epigenetic regulator. This layer of environmental regulation would be critical to acknowledge when investigating candidate genes implicated in BP regulation, and could in part explain difficulties in attempting to reproduce gene association studies. It could also explain inconsistencies in the literature regarding the variable BP response to salt intake and its role in human HTN. Finally, these findings, together with our previous report4, suggest that the LSD1+/−mouse may serve as a model for salt-sensitivity in human HTN (Table 3); however, the mechanisms responsible for this effect are yet to be determined
Table III.
Comparison of Phenotypes of LSD1 Gene Variants in Humans and LSD1+/−Mice
| Human studies | Animal studies |
|---|---|
Polymorphic Variants of LSD1 in African Americans:
|
Heterozygous LSD1 Knockout Mice:
|
HS=liberal sodium; LS=low sodium; PRA=plasma renin activity
The contribution of dietary salt to the pathogenesis of HTN has been an area of longstanding interest. Salt-sensitivity of BP is commonly present in essential HTN, particularly in African-Americans 3, 11–14. The pathophysiologic mechanisms leading to salt-sensitivity are complex and only partially understood. Our results, from animal studies and two hypertensive cohorts of different ethnicities, support LSD-1’s role as an epigenetic mediator of dietary sodium’s effect on BP. Interestingly, we did not observe this in the Caucasian cohort, despite an identical study protocol. This is unlikely to be explained solely by a lower MAF in the Caucasians (18% versus 28%) given the larger sample size, and may suggest a relatively intact dietary salt-LSD-1 relationship and/or compensatory mechanisms operating to dampen penetrance in Caucasians.
The underlying molecular pathways linking LSD-1 to salt-sensitive HTN remain to be elucidated but several possibilities exist. First, our exploratory studies suggest LSD-1’s effect is not due to dysfunctional RAAS activity. Increased AngII/RAAS activation have been shown to play a role in several types of BP salt-sensitivity1,15,19. However, data presented herein could not be explained by an activated RAAS, but rather volume expansion – induced thru an alternate mechanism – followed by “reactive” RAAS suppression. In the face of inappropriate salt retention, the RAAS should be quiescent. Indeed, LSD-1+/− mice displayed lower aldosterone levels than wild-type littermates4. Similarly, LSD-1 minor allele carriers in the African-American population had significantly lower circulating aldosterone. PRA was also reduced, but not significant perhaps due to assay sensitivity and small sample size. Second, aldosterone regulates the alpha subunit of ENaC24 to modify renal tubular sodium resorption. As would be anticipated with reduced aldosterone, LSD-1+/− mice displayed reduced alpha-ENaC expression. Whether this is due to decreased synthesis or increased catabolism is unknown. Third, our preliminary analyses suggest a possible abnormality in the renovascular response to dietary salt. Normally, when dietary sodium is increased, RBF increases to promote sodium excretion and maintain volume homeostasis. African-American minor allele carriers displayed no change in RBF, whereas African-American major allele homozygotes displayed an expected increase in RBF with HS intake. Thus, LSD-1 may influence renal vascular function by affecting the relationship between dietary salt, the RAAS and vascular function. Sodium excretion studies would be useful to confirm this possibility. Fourth, soluble guanylate cyclase, which seems to mediate in part the vascular phenotype in the LSD-1+/− mice is another potential candidate4. Fifth, protein kinase C activity has been suggested by Metzger et al25 to prevent LSD-1 from demethylating the H3K4 residue. Studies are needed to determine whether these systems interact with LSD-1 to induce volume expansion in vivo.
This study has several limitations. We assume, because of similar phenotypes between human minor allele carriers and LSD1+/− mice, the minor allele is associated with decreased LSD-1 levels and/or function in humans, although this was not specifically documented. The sample sizes of our human cohorts are small by genome-wide association study (GWAS) standards; however this is, a candidate gene study defined by work in the mouse. In addition, our strict phenotyping protocol reduces signal variance from potent confounders of vascular and hormonal responses (medication, posture, dietary salt, diurnal variation) enabling greater power to detect small changes with smaller sample sizes. This is further supported by findings in separate populations and concordance in a specific animal model. It is important to emphasize that LSD-1 tagging-SNPs identified as associated with salt-sensitive HTN are not necessarily causal. Similarly, LSD-1 is an epigenetic regulator of gene transcription. We have not identified other pathways (e.g. natriuretic peptide system, adrenergic system) that may be transcriptionally modified by LSD-1 and also mediate LSD-1’s role in the salt-sensitivity phenotype.
In summary, our findings suggest an interaction between dietary salt, LSD-1 and BP. Genetic variation in LSD-1 is associated with salt-sensitivity of BP. Further characterization of how LSD-1 affects genes involved in volume/pressure homeostasis is needed, but available data suggest that these genes are likely involved in vascular adaptation to changes in sodium intake. Thus, manipulation of LSD-1 activity may provide entrée to a novel paradigm for understanding the mechanisms underlying and/or treatment of salt-sensitive HTN.
Acknowledgments
We gratefully acknowledge the support of the dietary, nursing, administrative, and laboratory staff of the Clinical Research Centers in which these studies were performed. We also acknowledge the investigators, fellows, nurses and research coordinators at each of the study sites for the HyperPATH and HTN-IR study groups for their contribution to the study of these subjects. This research was supported by the following grants: The National Institutes of Health (NIH) grants HL47651, HL59424, DK63214, HL094452, HL67974, a Specialized Center of Research (SCOR) in Molecular Genetics of Hypertension (P50HL055000), P50-HL55005 (HTN-IR cohort), M01-RR000425 (Cedars-Sinai Medical Center), M01-RR000043 (University of Southern California), i2b2, a National Center for Biomedical Computing (NCBC) grant (LM008748) and Harvard Catalyst 5 UL1 RR025758. Dr. Chamarthi was supported by T32 HL007609 (NIH) and 5 KL2 RR025757 (Harvard Catalyst/NIH). Dr. JS Williams was supported by NIH grant K23 HL084236. Dr. Goodarzi was supported by R01 DK079888 and the Winnick Clinical Scholars Award. Dr. Pojoga was supported by KL2 RR025757 (NIH) and 0735609T (AHA Scientist Development Grant). Dr. Rotter was supported by the Cedars-Sinai Board of Governors’ Chair in Medical Genetics.
Footnotes
Disclosure: Authors have no conflicts of interest to declare
References
- 1.Williams GH, Hollenberg NK. Sodium-sensitive essential hypertension: emerging insights into an old entity. J Am Coll Nutr. 1989 Dec;8(6):490–494. doi: 10.1080/07315724.1989.10720318. [DOI] [PubMed] [Google Scholar]
- 2.Campese VM. Salt sensitivity in hypertension. Renal and cardiovascular implications. Hypertension. 1994 Apr;23(4):531–550. doi: 10.1161/01.hyp.23.4.531. [DOI] [PubMed] [Google Scholar]
- 3.Weir MR. Salt intake and hypertensive renal injury in African-Americans. A therapeutic perspective. Am J Hypertens. 1995 Jun;8(6):635–644. doi: 10.1016/0895-7061(95)00048-T. [DOI] [PubMed] [Google Scholar]
- 4.Pojoga LH, Williams JS, Yao TM, Kumar A, Raffetto JD, do Nascimento GR, Reslan OM, Adler GK, Williams GH, Shi Y, Khalil RA. Histone Demethylase LSD1 Deficiency during High Salt Diet is Associated with Enhanced Vascular Contraction, Altered NO-cGMP Relaxation Pathway, and Hypertension. Am J Physiol Heart Circ Physiol. Aug 26; doi: 10.1152/ajpheart.00513.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004 Dec 29;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005 Sep 16;19(6):857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, Liu F, Taylor H, Lozach J, Jayes FL, Korach KS, Glass CK, Fu XD, Rosenfeld MG. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007 Apr 19;446(7138):882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- 8.Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008 Jun;20(3):316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005 Sep 15;437(7057):436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, Rose DW, Glass CK, Fu XD, Rosenfeld MG. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007 Feb 9;128(3):505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campese VM, Parise M, Karubian F, Bigazzi R. Abnormal renal hemodynamics in black salt-sensitive patients with hypertension. Hypertension. 1991 Dec;18(6):805–812. doi: 10.1161/01.hyp.18.6.805. [DOI] [PubMed] [Google Scholar]
- 12.Nesbitt SD. Hypertension in black patients: special issues and considerations. Curr Hypertens Rep. 2005 Aug;7(4):244–248. doi: 10.1007/s11906-005-0020-5. [DOI] [PubMed] [Google Scholar]
- 13.Weinberger MH. Racial differences in renal sodium excretion: relationship to hypertension. Am J Kidney Dis. 1993 Apr;21(4 Suppl 1):41–45. doi: 10.1016/0272-6386(93)70074-9. [DOI] [PubMed] [Google Scholar]
- 14.Calhoun DA, Oparil S. Racial differences in the pathogenesis of hypertension. Am J Med Sci. 1995 Dec;310( Suppl 1):S86–90. doi: 10.1097/00000441-199512000-00016. [DOI] [PubMed] [Google Scholar]
- 15.Chamarthi B, Williams JS, Williams GH. A mechanism for salt-sensitive hypertension: abnormal dietary sodium-mediated vascular response to angiotensin-II. J Hypertens. May;28(5):1020–1026. doi: 10.1097/HJH.0b013e3283375974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang AH, Azen SP, Raffel LJ, Tan S, Cheng LS, Diaz J, Toscano E, Henderson PC, Hodis HN, Hsueh WA, Rotter JI, Buchanan TA. Evidence for joint genetic control of insulin sensitivity and systolic blood pressure in hispanic families with a hypertensive proband. Circulation. 2001 Jan 2;103(1):78–83. doi: 10.1161/01.cir.103.1.78. [DOI] [PubMed] [Google Scholar]
- 17.Williams JSY, Adler GK, Khalil RA, Romero JR, Sinha S, Ponnuchamy B, Williams GH. Genetic alteration of a histone demethylase is associated with altered aldosterone and vascular responsiveness: An intermediate phenotype of human hypertension. Hypertension. 2007;52:E38. [Google Scholar]
- 18.Chamarthi B, Kolatkar NS, Hunt SC, Williams JS, Seely EW, Brown NJ, Murphey LJ, Jeunemaitre X, Williams GH. Urinary free cortisol: an intermediate phenotype and a potential genetic marker for a salt-resistant subset of essential hypertension. J Clin Endocrinol Metab. 2007 Apr;92(4):1340–1346. doi: 10.1210/jc.2006-2093. [DOI] [PubMed] [Google Scholar]
- 19.Hopkins PN, Lifton RP, Hollenberg NK, Jeunemaitre X, Hallouin MC, Skuppin J, Williams CS, Dluhy RG, Lalouel JM, Williams RR, Williams GH. Blunted renal vascular response to angiotensin II is associated with a common variant of the angiotensinogen gene and obesity. J Hypertens. 1996 Feb;14(2):199–207. doi: 10.1097/00004872-199602000-00008. [DOI] [PubMed] [Google Scholar]
- 20.Weinberger MH. Salt sensitivity of blood pressure in humans. Hypertension. 1996 Mar;27(3 Pt 2):481–490. doi: 10.1161/01.hyp.27.3.481. [DOI] [PubMed] [Google Scholar]
- 21.Underwood RH, Williams GH. The simultaneous measurement of aldosterone, cortisol, and corticosterone in human peripheral plasma by displacement analysis. J Lab Clin Med. 1972 May;79(5):848–862. [PubMed] [Google Scholar]
- 22.Emanuel RL, Cain JP, Williams GH. Double antibody radioimmunoassay of renin activity and angiotensin II in human peripheral plasma. J Lab Clin Med. 1973 Apr;81(4):632–640. [PubMed] [Google Scholar]
- 23.de Bakker PI, Burtt NP, Graham RR, Guiducci C, Yelensky R, Drake JA, Bersaglieri T, Penney KL, Butler J, Young S, Onofrio RC, Lyon HN, Stram DO, Haiman CA, Freedman ML, Zhu X, Cooper R, Groop L, Kolonel LN, Henderson BE, Daly MJ, Hirschhorn JN, Altshuler D. Transferability of tag SNPs in genetic association studies in multiple populations. Nat Genet. 2006 Nov;38(11):1298–1303. doi: 10.1038/ng1899. [DOI] [PubMed] [Google Scholar]
- 24.Masilamani S, Kim GH, Mitchell C, Wade JB, Knepper MA. Aldosterone-mediated regulation of ENaC alpha, beta, and gamma subunit proteins in rat kidney. J Clin Invest. 1999 Oct;104(7):R19–23. doi: 10.1172/JCI7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metzger E, Imhof A, Patel D, Kahl P, Hoffmeyer K, Friedrichs N, Müller JM, Greschik H, Kirfel J, Ji S, Kunowska N, Beisenherz-Huss C, Günther T, Buettner R, Schüle R. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature. 2010 Apr 1;464(7289):792–6. doi: 10.1038/nature08839. [DOI] [PubMed] [Google Scholar]