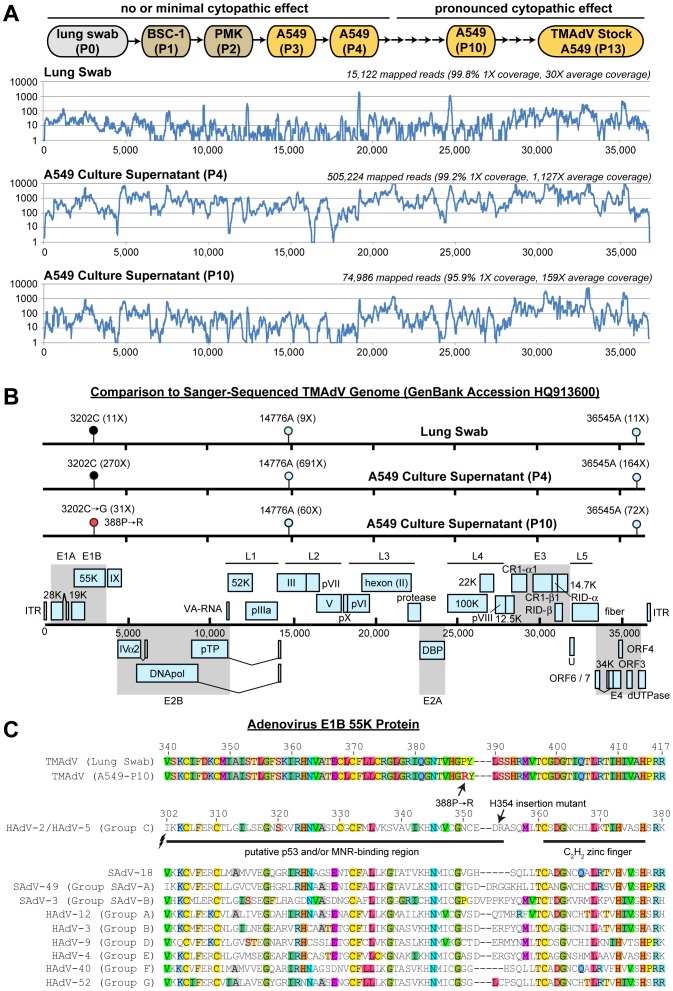
Figure 1. Whole-Genome Sequencing of Serially Passaged TMAdV.
(A) Genomic coverage of TMAdV by mapping of deep sequencing reads from a lung swab from a titi monkey with TMAdV pneumonia, passage 4 in A549 cells, and passage 10 in A549 cells. The coverage (y-axis) achieved at each position along the ∼37 kB TMAdV genome (x-axis) is plotted on a logarithmic scale. (B) Identification of single nucleotide polymorphisms (SNPs) in assembled TMAdV genomes. The coverage at each SNP position is shown in parentheses. SNP positions 14776A and 36545A (cyan lollipops) correspond to probable sequencing errors in the Sanger sequenced TMAdV genome (Genbank HQ913600). A 3202CG mutation is observed to occur between passage 4 (black lollipop) and passage 10 (red lollipop), which results in a 388PR amino acid coding change in the TMAdV E1B-55K protein. (C) A multiple sequence alignment of the E1B-55K protein of TMAdV and representative human and simian adenoviruses reveals that position 388 resides within a putative binding region for p53 and/or the MRE11-RAD50-NBS1 (MRN) complex. Human adenovirus type 5 (HAdV-5) mutants containing a nearby H534 insertion or mutations in the downstream C2H2 zinc finger inhibit binding to p53 and/or MRN [18].