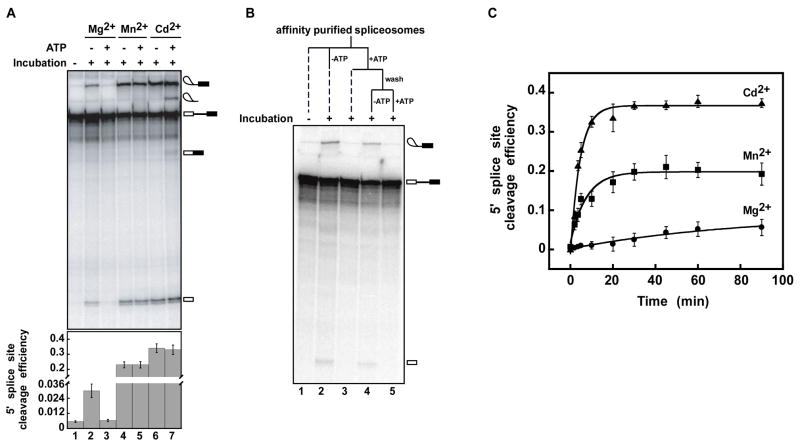
Figure 1. An ATP-dependent activity rejects pre-mRNA reversibly prior to 5′ splice site cleavage, if the rate of 5′ splice site cleavage is slow.
(A) An ATP-dependent activity competes with 5′ splice site cleavage. U6/sU80 (Sp) spliceosomes stalled in Mg2+ on an ACT1 pre-mRNA substrate were affinity-purified and then either frozen (no incubation) or incubated with the indicated metals and with or without ATP-Mg2+. The migration of the splicing species is indicated on the right. The bottom panel shows the efficiency of 5′ splice site cleavage. Values are represented as means ± standard error of mean (SEM) from three experiments. (See also Figure S1).
(B) ATP-dependent rejection is reversible. Spliceosomes were stalled and purified as in A and then either frozen (no incubation) or incubated −/+ ATP. Spliceosomes incubated with ATP were then washed to remove ATP and incubated further −/+ ATP. Dashed lines indicate that samples were not incubated further.
(C) In the absence of ATP, U6/sU80 (Sp) spliceosomes catalyzed 5′ splice site cleavage in Mg2+ but with an apparent rate 10-fold lower than that in Mn2+ and 20 fold lower than that in Cd2+ (kMg = 0.010 ± 0.006 min−1; kMn = 0.13 ± 0.02 min−1; kCd = 0.22 ± 0.02 min−1 – errors reflect the error in the fit of the data points (see Experimental Procedures)). Reactions were as in A but without ATP. Error bars represent ± SEM of each data point for three experiments.