Abstract
The G2/M arrest function of human papillomavirus (HPV) E4 proteins is hypothesized to be necessary for viral genome amplification. Full-length HPV18 E1^E4 protein is essential for efficient viral genome amplification. Here we identify key determinants within a CDK-bipartite consensus recognition motif in HPV18 E1^E4 that are critical for association with active CDK-cyclin complexes and in vitro phosphorylation at the predicted CDK phosphorylation site (threonine 23). The optimal cyclin-binding sequence (43RRLL46) within this E4 motif is required for G2/M arrest of primary keratinocytes and correlates with cytoplasmic retention of cyclin B1, but not cyclin A. Disruption of this motif in the E4 ORF of HPV18 genomes, and the subsequent generation of stable cell lines in primary keratinocytes revealed that this motif was not essential for viral genome amplification or L1 capsid protein induction. We conclude that the HPV18 E4 G2/M arrest function does not play a role in early vegetative events.
INTRODUCTION
Human papillomaviruses (HPV) are small double-stranded DNA viruses that infect cutaneous and mucosal epithelial tissues at many different body sites, gaining entry through micro-abrasions, and frequently inducing benign warts or papillomas. In most instances infections are self-limiting and the papillomas regress within a short time period, but some infections can occasionally persist and progress to cancer. Cancers associated with HPV are those formed in the anogenital tract and oropharynx, as well as some skin cancers. Whilst in excess of 100 HPV types have been identified, only 12 are deemed to be carcinogenic (Bouvard et al., 2009), with HPV16 and HPV18 having the greatest oncogenic risk (Bosch et al., 1995).
The HPV life cycle is intimately linked to the differentiation programme of the epithelium, with the virus first infecting cells of the basal layer. Establishment and maintenance of the viral genome in proliferating basal keratinocytes is followed by vegetative viral DNA amplification in cells that have moved up from the basal layer and differentiated. In normal epithelia, cells exit the cell cycle upon differentiation, but as the virus is dependent on the cellular replication machinery for the synthesis of its own DNA the virus induces these cells to re-enter the cell cycle in order to gain access to host replication factors (Hebner and Laimins, 2006). One of the most abundant HPV proteins produced during the infectious cycle is E4, a protein translated from spliced mRNA transcripts as an E1^E4 fusion protein and contains the first five residues of E1 fused to the remainder of E4 (Doorbar et al., 1986; Nasseri et al., 1987). The switch from maintenance replication of the viral genome to vegetative genome amplification is associated with induction of high levels of the E4 protein, with levels of E4 remaining high until completion of the HPV infectious cycle in the superficial cell layers (Peh et al., 2002).
The role of E4 within the virus infectious cycle is not under stood but a genetic analysis of E4 in keratinocyte-based model systems of HPV replication has shown a requirement for full-length E1^E4 expression in differentiation-dependent virus vegetative functions, including viral genome amplification of HPV types 16, 18 and 31 (Nakahara et al., 2005; Wilson, Fehrmann, and Laimins, 2005; Wilson et al., 2007). Disruption of the vegetative cycle is also a feature of rabbit papillomas induced by mutant cottontail rabbit papillomavirus genomes which do not support E1^E4 expression (Peh et al., 2004). Whilst these studies promote an essential role for E4 in the vegetative stages of HPV replication, this might not be the case for all viruses since E1^E4 did not have a critical role in HPV11 genome amplification (Fang et al., 2006).
Elucidation of E4 functions has largely been derived from studies of ectopic E4 expression systems and they have shown E4 to have diverse effects on cell behaviour and on cellular organization. These include the suppression of cellular DNA synthesis through inhibition of chromosomal replication origin licensing (Knight et al., 2004; Roberts et al., 2008), the inhibition of the G2-to-Mtransition of the cell cycle (Davy et al., 2002; Knight et al., 2004; Nakahara et al., 2002), and the promotion of apoptosis by alteration of mitochondrial function (Raj et al., 2004). The viral protein can have a destabilizing effect upon the keratin cytoskeleton (Doorbar et al., 1991; Roberts et al., 1993; Roberts et al., 1997) and on the cornified envelope (Bryan and Brown, 2000), and also induces the redistribution of the promyelocytic leukaemia protein from sub-nuclear domains (Roberts et al., 2003). Few interactions between E4 and host cell factors have been identified, but E4 proteins from different HPV types associate with the serine arginine kinase SRPK1; a kinase that regulates the function of mRNA splicing factors (Bell, Martin, and Roberts, 2007). Whilst together, these studies have promoted the idea of E4 as a multifunctional protein, the physiological significance of the various E4 functions to the role of E4 in HPV life cycle is not known.
The inhibition of G2-to-Mtransition is a function conserved between E4 proteins of diverse HPV types (HPV1, 11, 16 and 18), although investigations to date have shown that the various E4 proteins implement different strategies to inhibit cell division (Davy et al., 2006; Davy et al., 2005; Knight et al., 2004; Knight, Turnell, and Roberts, 2006; Nakahara et al., 2002). Since papillomavirus replication is dependent upon the host cell for provision of replication factors and enzymes, the stimulation of replication-activated cells to arrest in G2/M could have various benefits to the virus (Wang et al., 2009a). It could stimulate maintenance of a pseudo S phase-like state of the infected cell and indeed, cell cultures that have been induced to arrest in G2/M by E4 show evidence of continuous rereplication of cellular DNA (Knight et al., 2004; Nakahara et al., 2002). However cellular chromosomal rereplication is not always a consequence of E4 induced G2/M arrest and in some instances cellular DNA synthesis is inhibited (Knight et al., 2004; Roberts et al., 2008). An E4 mediated G2/M arrest of the cell cycle in replication-activated cells would therefore possibly allow the virus to access the host replication machinery without competing cellular DNA synthesis (Davy et al., 2002; Knight et al., 2004).
Therefore, to test the physiological significance of the E4 G2/M function within the HPV life cycle we have identified the sequence determinants of HPV18 E1^E4 necessary for induction of G2/M arrest in the host cells of the virus, primary human keratinocytes. HPV18 genomes containing mutations at these sites within the E4 open-reading frame (ORF) were generated and the ability of these genomes to support the vegetative life cycle in HFK was investigated.
RESULTS
Cytoplasmic retention of cyclins A and B1 by HPV18 E1^E4 in epithelial cells is dependent upon an RXL tripeptide
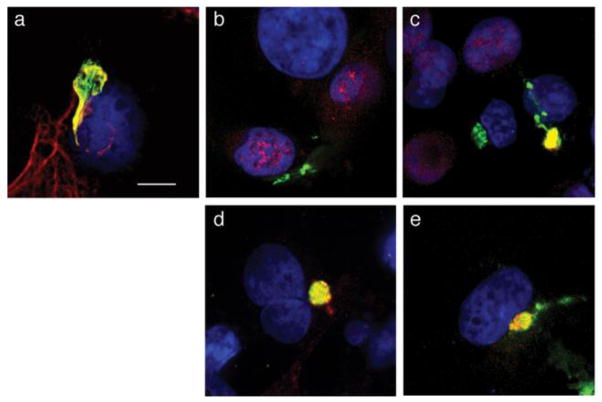
The mechanism of inhibition of mitotic entry by E1^E4 proteins of HPV11 and HPV16 involves pr event ion of nuclear shuttling of CDK1-cyclin B1 complexes, by tethering the active complexes to collapsed cytokeratin networks in epithelial cells (Davy et al., 2005). The HPV16 E4 arrest mechanism may also involveretention of the S-phase specific CDK2-cyclin A (Davy et al., 2006). It is likely that a similar mechanism under pins the induction of G2/M arrest by the related alpha virus HPV18 (Nakahara et al., 2002) and therefore we have examined whether HPV18 E1^E4 sequesters CDK-cyclin complexes to the cytoplasm of epithelial cells. In order to overcome poor expression of the native HPV18 E1^E4 cDNA in epithelial cells following transfection of plasmid DNA (our unpublished data), we switched most of the codons within the E1^E4 gene to those of the most frequently used codons found within the protein coding sequences of the human genome; a strategy that has been used to facilitate efficient synthesis of the HPV genes L1 and L2 in heterologous expression systems. Whilst this strategy replaces infrequently-used codons it can also disrupt negative regulatory elements in the mRNAs that might preclude efficient expression (Hindmarsh and Laimins, 2007; Zhou et al., 1999). Upon transfection of the codon-optimized (co) HPV18 E1^E4 cDNA into COS-1 epithelial cells, efficient expression of the protein is observed in at least 40% of cells, with E1^E4 located as a filamentous network radiating throughout the cytoplasm, or collapsed into juxtanuclear bundles. Co-staining with keratin 18 antibodies confirmed localization of E1^E4 to the keratin cytoskeleton (Fig. 1a), and staining with antibodies that recognize the different cyclin proteins revealed that cyclins A and B1, but not cyclin E co-localized with the cytoplasmic E4 structures (Fig. 1 b–d). Furthermore, the kinase partners of these cyclins, CDK1 and CDK2 also were co-localized with the E4 protein (Fig. 1e, and data not shown).
Figure 1. HPV18 E1^E4 colocalizes with CDK-cyclins in epithelial cells.
Confocal microscopy of COS-1 cells transfected with the plasmid expressing the codon-optimized HPV18 E1^E4 protein showed colocalization between HPV18 E1^E4 and keratin 18 (a), cyclin A (c), cyclin B1 (d), and CDK1 (e), but not cyclin E (b). Green, E4: Red, keratin 18, cyclin proteins and CDK1: Blue, DAPI-stained nuclei, and yellow indicates colocalization. Bar, 10 μm.
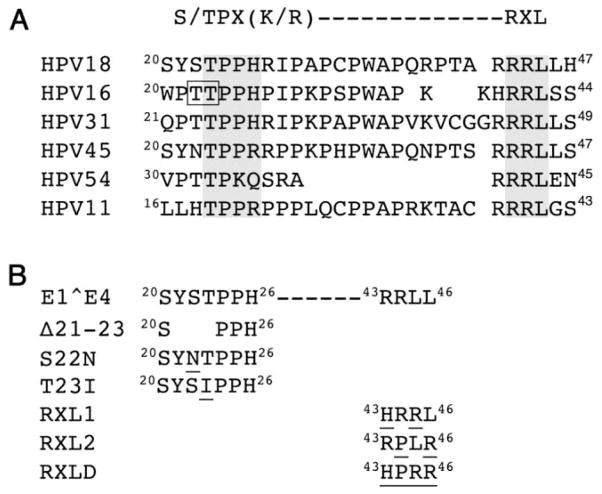
Proline-rich regions are a general feature of E1^E4 proteins and a threonine dipeptide (residues 22 and 23) within this region of the HPV16 E1^E4 protein is important for the cytoplasmic sequestration of cyclins A and B1 (Davy et al., 2006; Davy et al., 2005). A serine-threonine dipeptide (residues 22 and 23) in a corresponding proline-rich region of HPV18 E1^E4 might therefore serve an analogous role (Fig. 2A). However, deletion of residues 21 to 23 (E4Δ21-23, Fig. 2B) did not abrogate co-localization between E1^E4 and cyclin A (Fig. 3A) or cyclin B1 (data not shown) in the cytoplasm of COS-1 cells. Further examination of the HPV18 E1^E4 protein sequence identifies a CDK-bipartite consensus recognition motif (Takeda, Wohlschlegel, and Dutta, 2001), that consists of a CDK phosphorylation site (S/TPX[K/R], 23TPPH26) and a cyclin-binding sequence or Cy-motif (RXL,43RRL45). The recognition motif is a conserved feature in E1^E4 coding sequences of many, but not all HPV types in the alpha genus (Fig. 2A). Since we have already shown that loss of the putative CDK phosphoacceptor sitethreonine 23 does not affect E1^E4-mediated cytoplasmic sequestration of CDK-cyclin complexes we examined the contribution of the 43RRL45 sequence to this function. For some HPV types, including HPV18, there is a second overlapping RXL motif within this region of E1^E4 (44RLL46, Fig. 2A) and although this is not an optimal motif it may have cyclin-binding activity (Wohlschlegel et al., 2001). A series of mutations were therefore introduced into this region of HPV18 E1^E4 with amino acids selected that had the potential for maximum disruption of cyclin binding (Fig. 2B) (Wohlschlegel et al., 2001). The choice of amino acid substitutions that could be made however was limited due to a requirement to maintain the overlapping E2 coding sequence upon introduction of the mutations into the full-length HPV18 genome. Substitution at all positions within the 43RRLL46 sequences was made (HPRR; RXLD), as well as alteration of arginine 43 and leucine 45 in the consensus RXL sequence, 43RRL45 to form the tripeptide HRR (RXL1), and arginine 44 and leucine 46 were changed in the overlapping motif to form the tripeptide PLR (RXL2) (Fig. 2B). The mutagenic strategy however does not allow us to identify the relevance of each RXL tripeptide to CDK-cyclin sequestration since the overlapping nature of the RXL motifs makes it impossible to selectively modify one tripeptide without affecting the other.
Figure 2. E4 sequence contains a conserved bipartite CDK consensus recognition motif.
(A) Homology between a bipartite CDK consensus recognition motif and E4 coding sequences of HPV types of the alpha genus. Six examples are given, but other types within the genus also contain the motif within the E4 ORF. They are: HPV61, 72, 81, 83 (species 3); HPV18, 39, 45, 59, 68 (species 7); HPV43 (species 8); HPV16, 31 (species 9); HPV6, 11, 13 (species 10); HPV54 (species 13). The potential CDK phosphorylation sites (S/TPX[K/R]) and cyclin binding motifs (RXL) in E4 are identified by a grey-shaded box. A threonine dipeptide in HPV16 E1^E4 that is a key determinant in cytoplasmic retention of CDK-cyclin complexes is underlined (Davy et al., 2005). (B) Details of a deletion and substitutions made within the bipartite CDK consensus recognition sequence of HPV18 E1^E4; the substitutions are underlined.
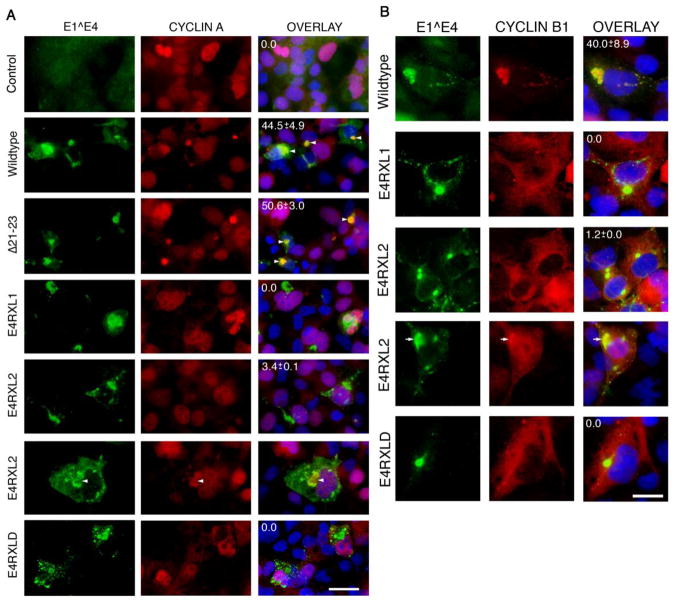
Figure 3. Disruption of RXL tripeptides in HPV18 E1^E4 abrogates cytoplasmic retention of cyclins A and B1.
(A) Immunofluorescence microscopy of COS-1 cells transfected with empty vector (control) or plasmids expressing HPV18 E1^E4 wildtype or the various mutants. Cells were co-stained with antibodies to E4 (green) and cyclin A (red), and nuclei counterstained with DAPI (blue). Cells showing sequestration of cyclin A to E4 inclusions is only evident in cells expressing the wild-type or deletion mutant (E4Δ21-23) E1^E4 proteins with examples identified with arrowheads. In a small sub-population of cells expressing the RXL2 mutation cyclin A was found to be associated with the collapsed E4-kerat in structures (arrowhead). Bar, 30 μm. (B) Staining of transfected COS cells for E4 (green) and cyclin B1 (red), and nuclei counter stained with DAPI (blue). In cells transfected with the plasmid expressing RXL2, rare cells are observed that show a degree of E4 and cyclin B1 co-staining (indicated by an arrow). Bar, 20 μm. The percentages of dual-positive cells (co-expressing E4 and cyclins A or B1) showing colocalization are given as means ± standard deviation from data derived from counting a minimum of 100 cells from three independent experiments.
Upon transfection into COS-1 cell s, all three RXL mutants were defective for cytoplasmic retention of cyclins A and B1 (Fig. 3), and CDK partners 1 and 2 (data not shown). However, the RXL2 mutant did retain a degree of co-localization with cyclins A and B1 but only in 3.5% (± 0.1) and 1.2% (± 0.0) of dual-positive cells respectively, markedly lower than the level of colocalization occurring in cells expressing the wildtype protein (44.5% ± 4.9 for E4 and cyclin A dual-positive cells and 40.0 ± 8.9 for E4 and cyclin B1 dual-positive cells) (Fig. 3). The efficiency of transfection with each plasmid was comparable (ranging between 36 and 41.5%) with the exception of the RXL1 plasmid that was consistently lower at ~ 30%. The steady-state expression levels of the RXL1 protein was also found to be consistently lower than the wildtype or the other mutant HPV18 E1^E4 proteins and suggests that the RXL1 mutation might have compromised the stability of the protein (data not shown).
The failure of these mutants to sequester CDK-cyclin complexes to the cytoplasm cannot be explained by interference with cytoskeleton reorganization since all three mutant proteins retained this biological property (data not shown).
The RXL motif of HPV18 E1^E4 mediates an association with active CDKs
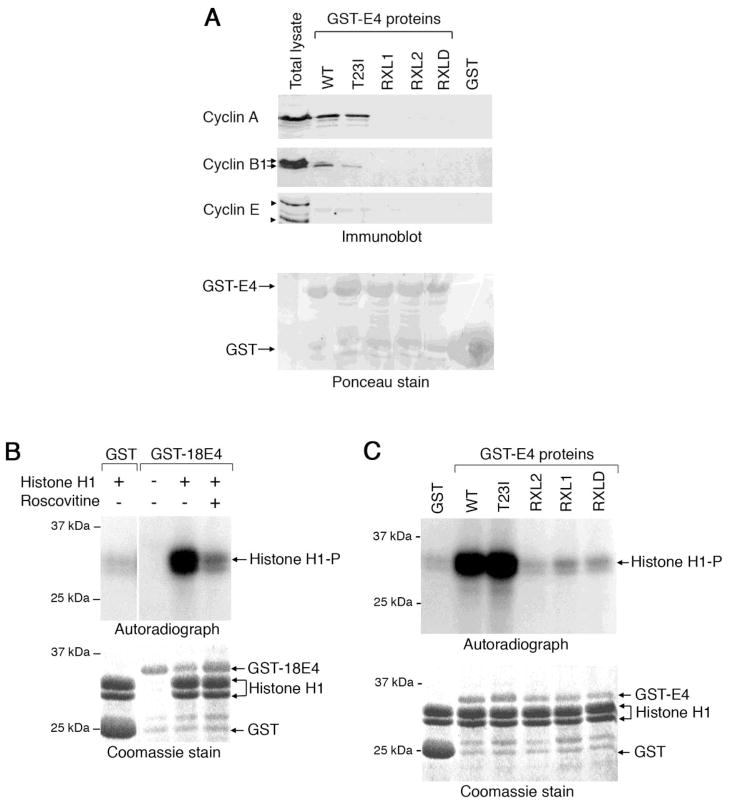
Since the 43RRLL46 sequence is necessary for the cytoplasmic retention of CDK-cyclin complexes we examined whether HPV18 E1^E4 can bind these cellular complexes via the RXL motifs. The poor solubility of the HPV18 E1^E4 protein in detergent-buffers prohibits co-immunoprecipitation experiments and therefore immobilized GST-HPV18 E1^E4 proteins, expressed and purified in bacteria, were used to co-precipitate cellular factors from an NP-40 extract of HeLa cervical tumour keratinocytes and the presence of cyclins E, A and B1 examined by Western blotting. The GST-E1^E4 protein was able to co-precipitate cyclin A, and to a lesser extent cyclin B1, but no interaction with cyclin E was observed (Fig. 4A). It is worth noting that the level of the slower migrating hyper-phosphorylated form of cyclin B1 that translocates to the nucleus (Toyoshima -Morimoto et al., 2001) was reduced in the GST-E1^E4 co-precipitates. Examination of the co-precipitations of the GST fusion proteins containing the series of mutations within the 43RRLL46 sequence revealed that none of the mutants were able to efficiently precipitate cyclin A or cyclin B1 (Fig. 4A).
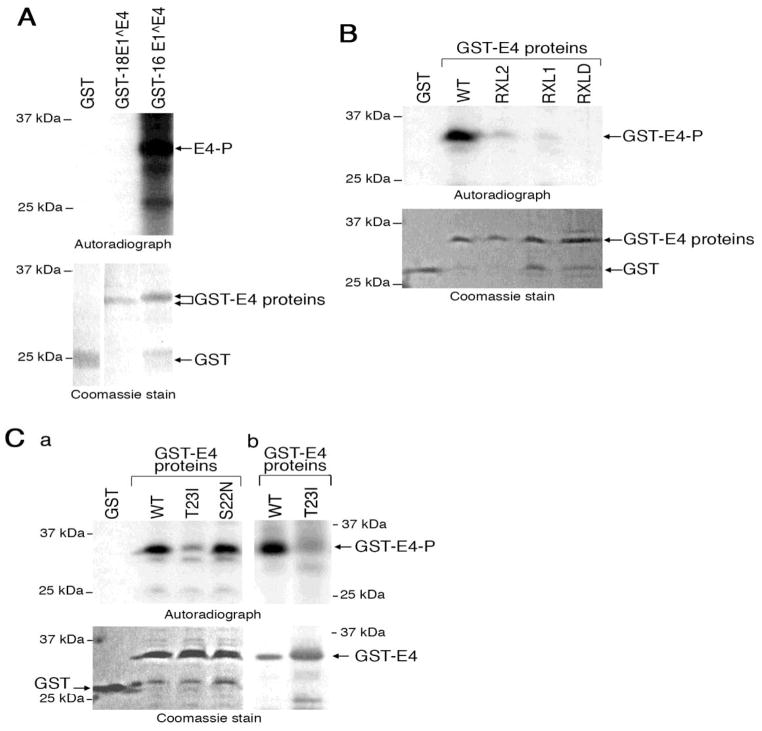
Figure 4. HPV18 E1^E4 association with active CDK-cyclin complexes is dependent on RXL motifs.
(A) The presence of cyclin A, B1 and E proteins in co-precipitates formed in a HeLa cell lysate using GST-fusion proteins of wild-type (WT) and mutant HPV18 E1^E4 proteins (immunoblot). Two forms of cyclin B1, indicated with arrows, are detected in the cell lysates, the slower migrating form being a phosphorylated form of the cyclin. Two forms of cyclin E (identified by arrowheads) were recognised by the antibody, the upper represents the mature form of cyclin E, ~ 47 kDa. Ponceau stain identifies loading of all of the GST-E4 proteins, with GST being in excess. (B) Phosphorylation of histone H1 protein by GST and GST-HPV18 E1^E4 co-precipitates prepared from HeLa cell lysates in the presence and absence of the CDK-inhibitor roscovitine (autoradiograph). The level of GST and histone H1 proteins is visualised by a Coomassie stain. (C) Phosphorylation of histone H1 by GST, wild-type (WT) and mutant GST-18E4 coprecipitates prepared from HeLa cell lysates. The Coomassie-stained gel shows loadings of the fusion proteins.
To establish whether HPV18 E1^E4 can bind to active CDK-cyclin complexes, co-precipitation assays from HeLa cell lysates were carried out using the different GST proteins and the level of associated CDK activity determined during an in vitro kinase assay using exogenous histone H1 as a substrate. Co-precipitates obtained using the HPV18 E1^E4 fusion protein mediated effective phosphorylation of histone H1, whilst addition of a GST protein co-precipitate to the in vitro reactions did not phosphorylate the histone substrate (Fig. 4B). To confirm that histone H1 phosphorylation by GST-HPV18 E1^E4-containing co-precipitations was CDK-dependent, the in vitro kinase reactions were performed in the presence of the CDK1-and CDK2-specific inhibitor roscovitine. The extent of histone H1 phosphorylation was reduced to near control levels, confirming that CDK1 and/or CDK2 associates with GST-HPV18 E1^E4 protein (Fig. 4B).
An assessment of the contribution of the RXL tripeptides to the association with CDK activity was investigated during the in vitro kinase assay using the mutant GST-E1^E4 proteins. The level of histone H1 phosphorylation in reactions containing co-precipitations of GST-E4 RXL1, RXL2 or RXLD was reduced to control levels (Fig. 4C). Taken together, our data indicates that the association between HPV18 E1^E4 and active CDK-cyclin complexes is dependent upon the integrity of the 43RRLL46 sequence.
CDKs phosphorylate HPV18 E1^E4 in vitro
Since the RXL mot if is common in CDK substrates we assessed whether E1^E4 is phosphorylated as a consequence of an association with active CDKs. HPV18 E1^E4 has only one consensus CDK recognition site, 23TPPR26 [S/TPX(R/K)], with threonine 23 acting as the phospho-acceptor site. This residue however is also a putative target of mitogen-activated protein kinase (p42 MAPK), as this region shares some identity with the consensus recognition sequence of this kinase (PXS/TP). To assess whether HPV18 E1^E4 is a substrate for these kinases we used an in vitro kinase assay using GST-HPV18 E1^E4 protein and purified forms of p42 MAPK and CDK2-cyclinA. In the presence of p42 MAPK, no phosphorylated bands were detected in the reactions containing GST or GST-HPV18 E1^E4 proteins, even though the kinase was able to phosphorylate a known p42 MAPK substrate, GST-HPV16 E1^E4 protein (Wang et al., 2009b) (Fig. 5A). In the CDK in vitro kinase assays however, a distinct phosphorylated species was observed in reactions containing the GST-HPV18 E1^E4 protein (Fig. 5B) indicating that the fusion protein is an in vitro substrate for CDK2-cyclinA, but not p42 MAPK.
Figure 5. CDKS phosphorylate HPV18 E1^E4 on threonine 23 in vitro.
(A) In vitro p42 MAP kinase phosphorylation assay of GST, GST-HPV16 and GSTHPV18 E1^E4. (B) In vitro phosphorylation of wild-type (WT) and mutant GST-18 E4 proteins by recombinant CDK2-cyclin A. (C) GST-HPV18 E4 containing substitutions at serine 22 (S22N) and threonine 23 (T23I) were used in an in vitro phosphorylation assay containing CDK2-cyclin A (a) and CDK1-cyclin A (b). The level of GST and GST fusion proteins in the reactions is shown in the Coomassie-stained gels.
Phosphorylation of the various GST-HPV18 E1^E4 RXL mutant proteins by CDK2-cyclin A was severely diminished (RXL1 and RXL2) or abrogated completely (RXLD) (Fig. 5B), suggesting that efficient phosphorylation of E1^E4 by CDK2-cyclin A requires physical interaction through the RXL tripeptides.
To identify the target residue of CDK2 within HPV18 E1^E4, the in vitro kinase assay was repeated in the absence of radioactive isotope and the GST fusion protein analysed by NL-ECD MS/MS, a technique for identification of sites of protein phosphorylation. An HPV18 E1^E4 phosphopeptide 12YPLLSLLNSYSTPPHR27 was identified with predicted phosphorylation at threonine 23. To confirm the results of the mass spectrometry analysis, in vitro CDK2 kinase assays were performed using GST-HPV18 E1^E4 proteins containing substitutions of threonine 23 (threonine 23 -> isoleucine, T23I) or of the adjacent serine residue serine 22 (serine 22 -> asparagine, S22N) (Fig. 2B). (In the context of the intact HPV18 genome, these are the only codon changes possible at these positions within the E4 sequence without altering the coding capacity of the E2 ORF.) The extent of phosphorylation of the S22N mutant protein was comparable to that of the wild-type protein (Fig. 5C). In contrast, in vitro phosphorylation of T23I was severely reduced, supporting the mass spectrometry data in the conclusion that threonine 23 in HPV18 E1^E4 is a CDK2-cyclin A phosphorylation site in vitro (Fig. 5C, a).
Purified CDK-cyclin kinases CDK2-cyclin E, CDK1-cyclin A and CDK1-cyclin B1 were also able to phosphorylate HPV18 E1^E4 in the in vitro assay and for all of these CDK-cyclin complexes, threonine 23 which is the sole consensus CDK phosphorylation site within the HPV18 E1^E4 sequence, the accept or site (data not shown and Fig. 5 C, b).
Whilst threonine 23 was identified as the phosphorylation site of CDK2, co-precipitation analysis using a GST-HPV18 E1^E4 protein containing the isoleucine substitution of threonine 23 demonstrated that this site was not critical for the association with active CDK-cyclin complexes (Fig. 4, A and C).
Inhibition of G2-to-M transition of primary foreskin keratinocytes by HPV18 E1^E4 is dependent on the integrity of the RXL tripeptides
Since HPV18 E1^E4, like the type 16 protein, sequesters CDK-cyclin complexes to the cytoplasm, this action is likely to be necessary for the HPV18 E1^E4 induced G2/M arrest of the cell cycle. To confirm this, HPV18 E1^E4 was expressed in primary human keratinocytes, the relevant host cell of the virus. The viral protein was expressed in human foreskin keratinocytes (HFK) cells using a recombinant adenovirus expressing the E1^E4 cDNA (r Ad18E1^E4). The infections were performed on cells isolated from two foreskin donors and a recombinant adenovirus expressing β-galactosidase was used to control for adenovirus effects on the cell cycle. At a multiplicity of infection of 50, the efficiency of infection was no less than 70% for both viruses; and examination of the cell cycle profiles after 36 h showed a marked increase in the G2/M population of rAd18E1^E4-infected cells, compared to mock-infected cells or to cells infected with the β-galactosidase-expressing virus (Fig. 6A).
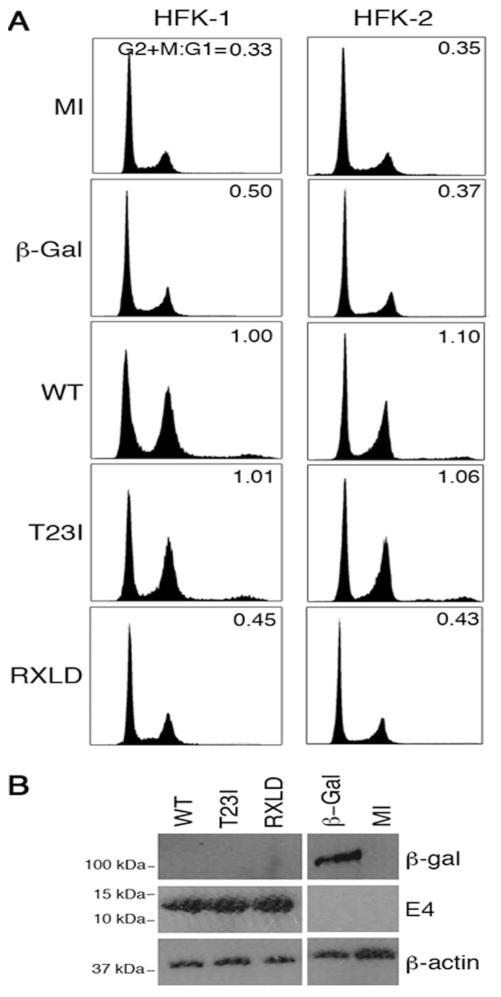
Figure 6. Integrity of RXL tripeptides is required for HPV18 E1^E4 induced G2/M arrest in primary foreskin keratinocytes.
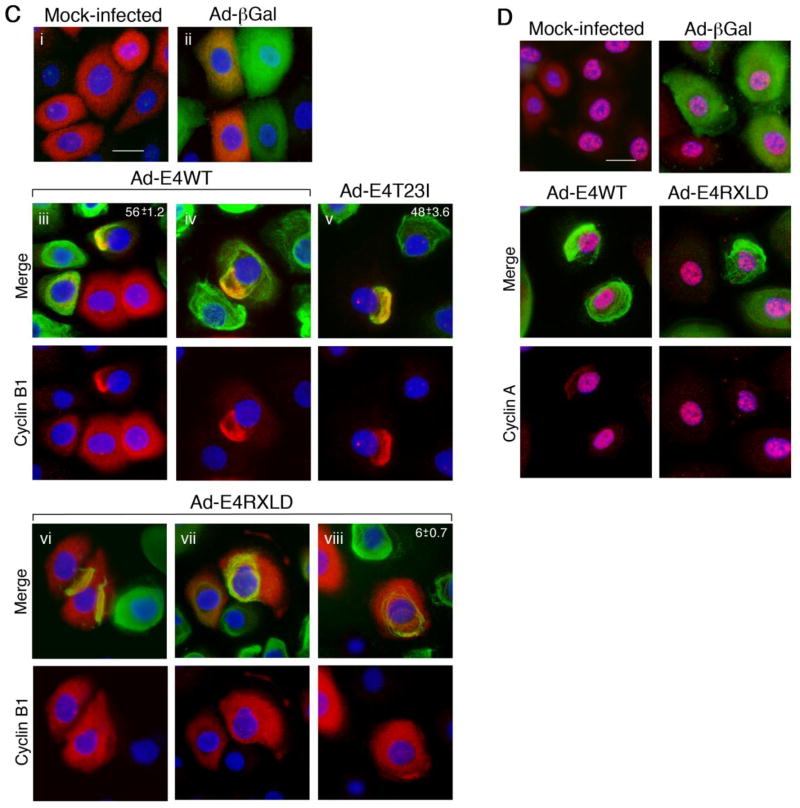
(A) Cell cycle profiles of primary human foreskin keratinocytes (HFK) infected with recombinant adenoviruses expressing β-galactosidase (βGal), HPV18 E1^E4 (WT) or the mutants T23I or RXLD for 36 h. The percentages of cells in G1 and in G2/M were used to calculate the G2+M:G1 ratios. Results are show for keratinocytes derived from two different donors and are representative of at least four independent experiments. (B) Western blot analysis of cell lysates showing the level of expression of the different proteins using β-actin levels as a loading control. (C) Cellular distribution of cyclin B1 (red) in HFK mock-infected (i), infected with the virus expressing (βGal) (ii), or with viruses expressing E4WT, E4T23I or E4RXLD (βGal and E4 proteins shown in green). Cyclin B1 is sequestered to both fully collapsed E4 filamentous networks (iii) as well as those that have undergone a partial collapse (iv). Mutation of threonine 23 (v) did not abrogate the association. Cyclin B1 was not sequestered to the collapsed filamentous networks of RXLD (vi and vii), although occasional cells showed a partial co-distribution (viii). Nuclear stain (DAPI) is shown in blue. (D) Cyclin A (red) distribtion in HFK, either mock-infected or infected with viruses expressing βGal, E4WT or E4RXLD proteins (all shown in green). Nuclear stain (DAPI) is shown in blue. Bar, 10 μm. The percentages of dual-positive cells is given as the means ± standard deviation, in the top-right of the panel s. Data was derived from three counts, each of a minimum of 100 cells.
To determine if the CDK–bipartite recognition motif of E4 was involved in the G2/M arrest function, two further recombinant viruses were generated, one expressing the mutant containing the substitution of the phospho-acceptor site (T23I), and the other protein with substitutions within the RXL tripeptides (RXLD). The cell cycle profiles of primary keratinocytes infected with these viruses clearly showed that disruption of the RXL motifs abrogated the G2/M arrest function whilst loss of the CDK phosphorylation site did not affect this E4 function (Fig. 6A). Western blot analysis confirmed that the E4 proteins were expressed at comparable levels to one another (Fig. 6B). We therefore conclude that disruption of the integrity of the sequence 43RRLL46 correlates with loss of the G2/M arrest function in primary keratinocytes, and thus represents a key determinant in cell cycle disruption by HPV18 E1^E4.
Immunofluorescence microscopy of the infected cells was used to establish if the loss of G2/M arrest of primary keratinocytes correlated with a failure of the E4 protein to sequester cyclins to the cytoplasm. Within HPV18 E1^E4-expressing HFK cultures, collapse of the E4 filamentous network into a juxtanuclear structure is less frequent than in transformed cells such as COS-1, and in a majority of cells the networks remain intact or only partially collapsed, even at late times post -infection. However, in these cells colocalization between cyclin B1 and cytoplasmic E4 was evident in 56% (± 1.2) of the co-stained cells (Fig. 6C). In contrast, in cells infected with the virus expressing the RXLD mutant protein, the majority of dual-positive cells did not show sequestration of cyclin B1 to RXLD cytoplasmic structures (Fig. 6C, Ad -E4RXLD panels vi and vii) and only 6% (± 0.7) of the dual -positive cells showed co-distribution between the two proteins, and this was only a partial colocalization (Fig. 6C, Ad -E4RXLD panel viii). Mutation of the threonine phospho-acceptor did not abrogate cytoplasmic sequestration of cyclin B1 with 48% (± 3.6) of the dual-positive cells showing colocalization (Fig. 6C). Examination of cyclin A distribution in HPV18 E1^E4-expressing cells revealed that the protein remained predominantly nuclear, even in cells in which the E4 filamentous network had collapsed (Fig. 6D). Within some of these cells however, the low level of cyclin A detected in the cytoplasm is sequestered to the collapsed E4 structures, and this is not observed in RXLD-expressing cells (Fig. 6D).
Genomes containing mutations within the CDK-bipartite recognition motif of E4 are established in primary foreskin keratinocytes
In our previous study we showed that a mutant HPV18 genome that contains a stop codon in the E4 sequence that terminates the E1^E4 protein at residue 16 (E4M17) and effectively precludes expression of the full-length E1^E4 protein, did not amplify efficiently upon induction of epithelial differentiation (Wilson et al., 2007). To test the hypothesis that the E4 G2/M arrest function is necessary to support virus genome amplification we inserted the RXLD mutation into the E4 ORF of complete HPV18 genomes. As mentioned, the mutations were designed to alter the coding sequence of E4, but not the integrity of the overlapping E2 coding sequence. Although threonine 23 does not play a role in G2/M arrest of HFK, we have shown it to be a phospho-acceptor site in CDK in vitro phosphorylation assays (Fig. 5), and hence it may contribute to E4 function in viral genome amplification. Therefore a mutant HPV18 genome containing the T23I mutation in the E4 ORF was also included in our analysis.
HFKs isolated from three different donors were transfected with the wild type or mutant HPV18 genomes to produce stable cell lines. Total genomic DNA was isolated from monolayer cultures of the different cells lines and Southern blot analysis used to identify the presence of HPV18 episomes. Significant levels of viral episomes were present in cells containing the wild type and mutant genomes indicating that mutation of the CDK phosphorylation site or the RXL sequences in the CDK-bipartite recognition motif of E4 do not inhibit episome establishment replication in HFKs (Fig. 7A). Whilst the level of episome copy number was similar between the different cell lines, in one of the three HFK isolates, the RXLD genomes were established at higher levels than the other genomes; however this effect was not observed in the two other donors (Fig. 7A and data not shown). None of the mutations within the CDK-bipartite recognition motif of E4 had any deleterious effect upon stable maintenance of the viral DNA upon extended passage of the different cell lines (data not shown).
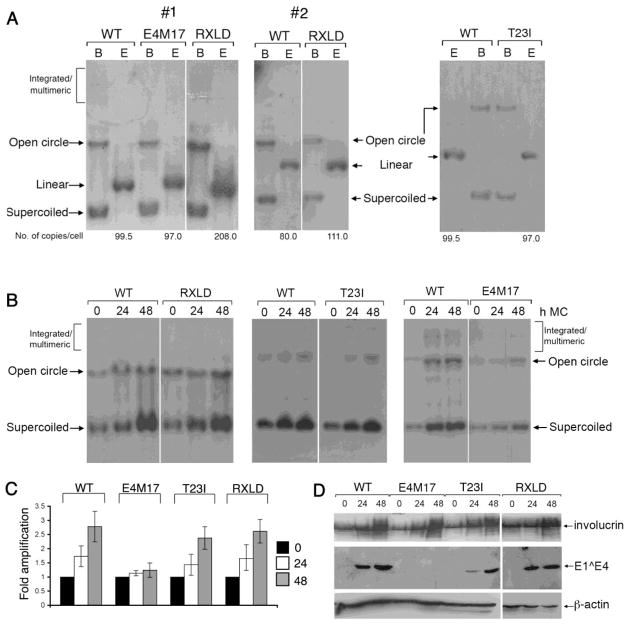
Figure 7. The CDK-bipartite recognition motif in E4 is not essential for establishment of the viral genome or differentiation-dependent viral genome amplification.
(A) Southern blot analysis of HFK stably transfected with HPV18 wild-type (WT), RXLD and E4M17 mutant genomes. Equal amounts of total DNA was digested with DpnI to remove residual input bacterial DNA and either BglII (B), which does not have a site in the HPV18 genome or EcoR1 (E), which has a single recognition site in the HPV18 genome and produces linear forms of the viral genome. HPV18 episomes migrate as supercoils and open circles. Whilst in one donor (#1), the RXLD genomes established at a higher copy number than the wild type genomes this was not a consistent observation across all donors (e.g. 2). (B) Southern analysis of equal amounts of total genomic DNA isolated from keratinocyte cell lines containing wild-type (WT) or mutant (T23I, RXLD, E4M17) HPV18 genomes grown in monolayer cell culture (0 h) or suspended in methylcellulose (MC) for 24 and 48 h. DNA was digested with DpnI and BglII and HPV18 episomes migrate as supercoils and open circles. (C) Bargraph of phosphoimager analysis of Southern blots to show levels of HPV18 DNA genome amplification. Data were derived from at least fifteen (WT and RXLD) or six (E4M17 and T23I) experiments of each cell line in a total of three different donor keratinocytes. (D) Western blot analysis of the total protein extracts isolated from the different HPV18 genome-containing cell lines to determine expression levels of HPV18 E1^E4 protein. The induction of differentiation was monitored by determination of the expression levels of the epidermal differentiation-marker involucrin. Levels of β-actin were used as a loading control.
We have previously shown that loss of expression of full-length E1^E4 (E4M17) was associated with an increase in the growth rate of the E4M17 mutant genome–containing cell lines compared to the wild type cells (Wilson et al., 2007). Similar analyses of the growth of the mutant genomes generated in this study showed no difference in rate of growth when compared to cells containing the wild type genomes (data not shown).
Differentiation-dependent HPV18 DNA amplification is not inhibited in keratinocytes containing mutations within the CDK-bipartite recognition motif of E4
Suspension of HPV genome containing keratinocytes in methylcellulose stimulates cellular differentiation and activation of viral genome amplification (Ruesch, Stubenrauch, and Laimins, 1998). To establish if the functions mediated by the HPV18 E4 CDK-bipartite recognition motif are important for this viral event, the HPV18 genomes containing the mutations within this motif were suspended in 1.5% methylcellulose for 24 and 48 h. Southern analysis of total genomic DNA was used to determine the level of HPV18 DNA amplification. Efficient amplification of viral episomes containing the RXLD mutation (2.6 ± 0.4 fold), and the T23I mutation (2.4 ± 0.4 fold) was observed in the three donor backgrounds, with both T23I and RXLD genomes amplifying to levels comparable to the wild type genomes (2.8 ± 0.5 fold, a level of amplification observed with other high risk types (Wilson, Fehrmann, and Laimins, 2005)) (Fig. 7 B and C). Consistent with our previous findings (Wilson et al., 2007), cells containing the E4 “knock-out ” mutant genome (E4M17) show poor amplification of the mutant genome upon suspension in methylcellulose (1.24 ± 0.25 fold). Amplification of the wild-type, T23I and RXLD genomes correlated with induction of late gene expression including high-levels of E1^E4 protein; and as expected E1^E4 protein could not be detected in differentiated cells containing the E4M17 genome (Wilson et al., 2007) (data not shown and Fig. 7D). The degree of differentiation induced in the cells lines carrying the various mutant genomes was monitored by expression of involucrin, a marker of differentiation (Fig. 7D). All the cell-lines, including E4M17 induced involucrin to levels comparable to those induced in cells containing the wildtype HPV18 genome.
HPV18 genomes containing mutations that disrupt the E4 RXL tripeptides support L1 capsid protein expression
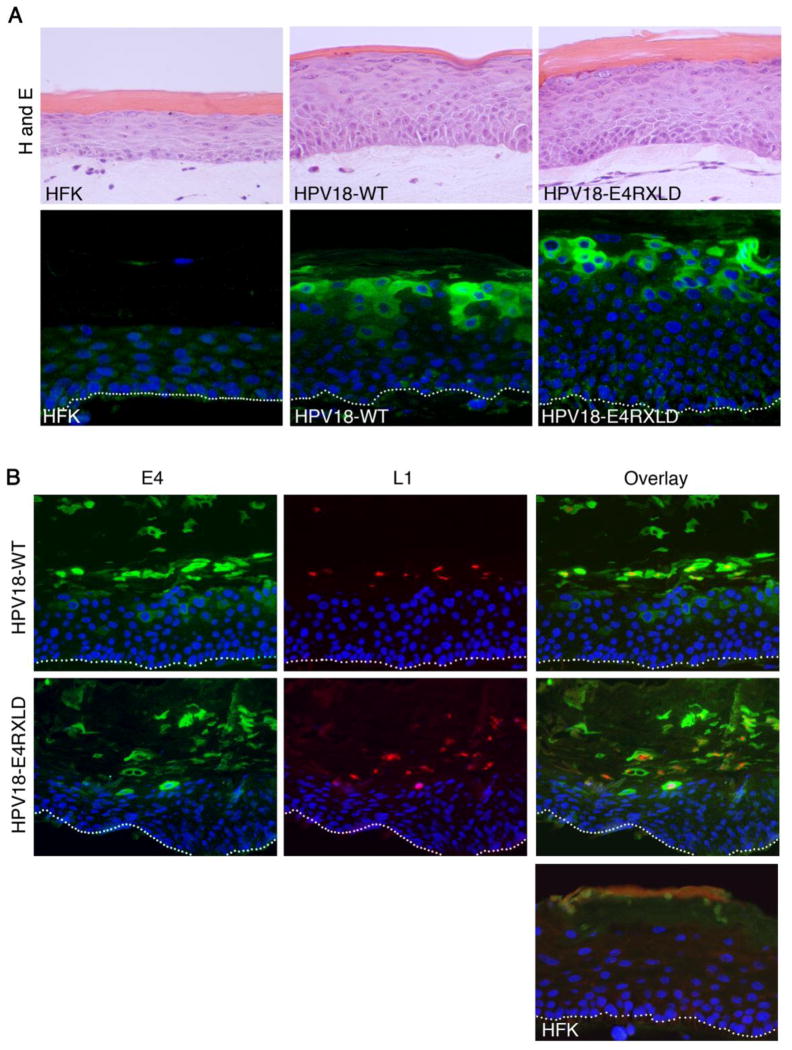
Whilst the integrity of the RXL tripeptides is important for G2/M arrest of HFK it is not necessary for efficient viral genome amplification. The function of this conserved E4 motif may however be of importance to other vegetative events. It has recently been shown that the papillomavirus L1 capsid gene is preferentially translated in keratinocytes when in a G2/M-like state (Ding et al., 2010), and since E4 protein is present in infected cells competent for capsid expression (Peh et al., 2002), perhaps the E4 G2/M arrest function is required for the expression of the capsid proteins. Thus, to examine whether cells expressing the HPV18 mutant genome defective in G2/M arrest (RXLD) supports L1 protein expression, the cells were stratified in organotypic raft culture. The stratified RXLD structures adopted the typical morphology associated with HPV genome containing cell s; thickening of the stratified layers compared to the rafts produced from the untransfected primary keratinocytes, and also retention of nuclei in the upper layers (Fig. 8A, H and E). Following staining with an E4 polyclonal antibody, abundant E4 protein is detected in the cytoplasm of cells of the upper stratified cell layers of both the wild type and RXLD rafts (Fig. 8A, E4). Examination of the co-distribution of E4 and L1 proteins in these rafts identified L1 expression in E4-positive cells of both wild type and RXLD rafts and indicates that superficial cells expressing the E4-RXLD protein are able to support expression of the L1 protein.
Figure 8. Disruption of the RXL tripeptides within E4 does not block expression of major capsid protein L1 in organotypic rafts.
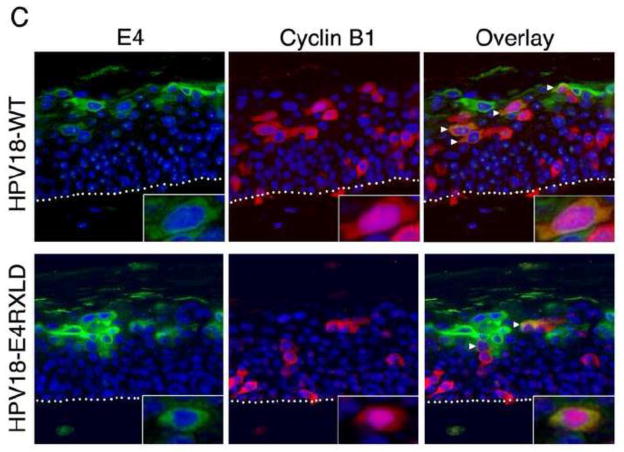
(A) Sections of organotypic rafts were stained with haematoxyl in and eosin (H and E) or immunohistochemistry performed for detection of E4 (green) and nuclei counterstained with DAPI (blue). (B) Sections stained for E4 (green), L1 (red) and counter stained with nuclear stain DAPI (blue). Areas of the rafts formed from cells containing HPV18 wild type genomes or the mutant RXLD showed expression of the capsid protein in E4-positive cells of the superficial cell layers. Rafts generated from donor keratinocytes (HFK) were negative for E4 and L1 expression. (C) Following co-staining of sections for E4 and cyclin B1, E4-positive cells containing cytoplasmic cyclin B1 are present in rafts generated from cells containing the wild-type or the RXLD genomes (arrowheads). Nuclear cyclin B1 is present in a few E4-positive cells in both wild-type and RXLD rafts (inset). Images of organotypic rafts are typical of staining observed between at least three organotypic rafts of each cell line.
The combined data of the analysis of the HPV18 genome-containing cell lines indicates that the function of the RXL motif within HPV18 E1^E4 does not play a significant role in vegetative events such as viral genome amplification or expression of the major capsid protein. This motif is a key determinant of E4-mediated G2/M arrest in HFK grown in monolayer culture, however, following stratification of the HPV18 wild-type genome-containing cells, few show colocalization between E4 and cytoplasmic cytoplasmic B1 (Fig. 8C), a finding that concurs with other studies of both HPV18 and HPV16 (Chow et al., 2009; Davy et al., 2005). Furthermore, cyclin B1 is maintained in the cytoplasm of cells expressing the RXLD mutant genomes (Fig. 8C), with dual-positive cells occurring at equivalent frequencies within the wild-type (3.18 ± 1.53) and RXLD (3.37 ± 0.88) (minimum of 300 E4-positive cells counted between three replicates) genome containing rafts. However, E4-RXLD positive cells containing nuclear cyclin B1 are occasionally detected, but nuclear cyclin B1 is also observed in stratified cells expressing the wild-type E4 protein (Fig. 8C; inset).
DISCUSSION
Coincidence between the onset of viral genome amplification and initiation of abundant production of E4 in natural infections is highly suggestive of a role for E4 in this vegetative phase of the virus life cycle (Breitburd, Croissant, and Orth, 1987; Peh et al., 2002). Indeed, inefficient genome amplification in papillomavirus-containing keratinocytes is one consequence of loss of full-length E1^E4 expression (Nakahara et al., 2005; Peh et al., 2004; Wilson, Fehrmann, and Laimins, 2005; Wilson et al., 2007). Induction of cells already returned to S phase by E7 to arrest in G2/M could stimulate maintenance of a pseudo S phase-like state of the infected cell, and indeed, cell cultures that have been induced to arrest in G2/M by E4 show evidence of continuous rereplication of cellular DNA (Knight et al., 2004; Nakahara et al., 2002). However cellular rereplication is not always a consequence of E4 induced G2/M arrest and in some instances cellular DNA synthesis is al so inhibited, by E4 blocking the formation of pre-replication complexes at the cellular origin of replication (Knight et al., 2004; Roberts et al., 2008). An E4 mediated G2/M arrest of the cell cycle in replication-activated cells may therefore inhibit competing replication of the host genome so that the virus can have exclusive access to the host’s biosynthetic machinery for high-level synthesis of the viral DNA (Davy et al., 2002; Knight et al., 2004). Evidence to indicate that HPV may not be able to amplify their genomes efficiently whilst the cell is actively replicating its own DNA has emerged from studies of organotypic raft culture of HPV16 and 18 genome-containing cells. In these cell -based models of the HPV replication cycle, viral genome amplification was observed to be largely confined to cells that show little evidence of cellular DNA synthesis (Nakahara et al., 2005; Wang et al., 2009a). In the HPV18-keratinocyte model, these cells contained high levels of cytoplasmic cyclin B1 and it was therefore proposed that HPV18 genome amplification is dependent on infected cells arresting in G2/M (Wang et al., 2009a). Our finding that the G2/M arrest function of HPV18 E1^E4 is not an essential function for efficient viral genome amplification in differentiated cells suggests that a G2/M environment is not necessary for HPV18 genome amplification, or if it is then factors other than E1^E4 are required to generate this cellular environment. The latter possibility might indeed be true since the cytoplasmic accumulation of cyclin B1 in the HPV18 organotypic rafts cultures preceded the detection of E1^E4 protein (Chow et al., 2009) (although it cannot be ruled out that in these cells E1^E4 is expressed but at a level that precludes detect ion with the E4 antibodies).
Whilst the HPV18 E4 G2/M function is not essential for viral genome amplification the translation of viral genes, and/or the assembly of specific protein complexes, necessary for efficient virion morphogenesis might be better facilitated in a G2/M environment (Andersen, Le Rouzic, and Planelles, 2008; Chulu et al., 2010). Indeed, translation of the L1 capsid protein has been shown to be more favourable in a G2/M environment (Ding et al., 2010). In our study however disruption of the 43RRLL46 motif within E4 does not lead to a marked reduction in late viral gene expression and cells expressing this mutant genome maintain expression of the L1 protein in superficial cells of organotypic rafts, indicating that the function of this motif is not critical for these early events in virion morphogenesis.
In primary cells expressing HPV18 E4, failure of these cells to arrest in G2/M was dependent on the integrity of the E4 cyclin–binding motif and correlated with loss of cytoplasmic colocalization between E4 and cyclin B1. Interestingly, the HPV1 E4 protein induces G2/M arrest using two different strategies, regulated by N-terminal processing of the viral protein (Knight et al., 2004; Knight, Turnell, and Roberts, 2006). Since HPV18 E1^E4 is also a substrate for N-terminal proteolysis in differentiating keratinocytes (A.G.P. and S. R., unpublished data) then it is conceivable that it might also encode multiple mechanisms of regulating cell division. An alternative E4-mediated G2/M arrest mechanism, other than cyclin B1 sequestration, active in replication–activated suprabasal keratinocytes could be an explanation of the infrequency of suprabasal cells showing colocalization between E4 and cyclin B1 in organotypic raft cultures of HPV genome-containing cells as observed by us, and others (Chow et al., 2009; Davy et al., 2005).
However, whilst our study shows that ectopic expression of HPV18 E4 in undifferentiated primary keratinocytes induces a G2/M arrest, it does not prove that this E4 function is active within replication-activated suprabasal cells; the site of abundant E4 expression in a natural HPV infection, and the possibility that a block in cell division is an artefact of over expression of E4 in undifferentiated proliferating cells cannot be excluded (Chow et al., 2009). It is worthwhile noting that a comparison of the cell cycle profiles of HPV18 containing keratinocytes following activation of differentiation-dependent viral DNA amplification did not identify any differences between the profiles of wild-type, E4-KO or RXLD genome containing cells (GLK and SR, unpublished data).
The conservation of the bipartite CDK binding motif in E4 proteins amongst the alpha group viruses indicates that it probably serves an important role in E4 function. Mutation of the putative CDK-phosphoacceptor residue (threonine 23) in the HPV16 protein was found to be critical for maintaining G2/M arrest in Schizosaccharomyces pombe, (although this proved not to be the target site of CDK2 phosphorylation (Davy et al., 2006)), but mutation of the RXL motif (40RRL42) did not hinder E1^E4 induced cell cycle arrest (Davy et al., 2002). Whilst the mechanism of G2/M arrest between these two related viruses (HPV16 and HPV18) appears to be linked to cytoplasmic sequestration of the mitosis-promoting factor there might however be variation in specific sequence requirements for its induction. It should be noted that the contribution of the RXL motif to HPV16 E4-mediated sequestration of CDK-cyclins in keratinocytes was not tested and moreover, discrepancies have been reported between the HPV16 G2-arrest mechanism in S. pombe and mammalian epithelial cells (Davy and Door bar, 2007).
In this study, sequestration of cyclin B1 to the cytoplasm was a common effect of over-expression of the type 18 E1^E4 protein in primary keratinocytes, and in SV40-transformed epithelial COS-1 cell s; and in both cell types this event is dependent on the integrity of the RXL motif. However unlike in COS-1 cell s, HPV18 E1^E4 does not induce cytoplasmic retention of cyclin A in primary cells, indicating that this E1^E4 function is not necessary for the G2/M arrest of primary cells. A specific modification of HPV18 E1^E4 that occurs in COS-1 cells but not HFK might be required for the interaction between E1^E4 and cyclin A, or alternatively the association is mediated by a factor not expressed in the monolayer cultures of primary foreskin cells. In fact, an unidentified factor has been implicated in mediating the interaction between HPV16 E1^E4 and cyclin A in cervical cancer cells, which is favoured in the G2 phase of the cell cycle (Davy et al., 2006). If cell type-specific E4 interactions do occur then perhaps the complete repertoire of E4 functions are not evident in the foreskin keratinocyte cell–based model used in our study. Further investigations are therefore necessary to determine the biological relevance of an association between HPV18 E1^E4 and CDK-cyclin A in HPV-infected cells.
CDK phosphorylation of the E1^E4 protein could modify E1^E4 behaviour during the infectious cycle. MAPK phosphorylation of HPV11 and 16 E1^E4 for example leads to changes in subcellular location of the viral protein, and the association between HPV16 E1^E4 and host keratins is enhanced (Bryan et al., 2000; Wang et al., 2009b). Only one S/TP site (23TP24) is present within HPV18 E1^E4 and at least in vitro it is the preferred phospho-acceptor site for kinases CDK1 and CDK2, and not p42 MAPK. Whilst there is no role for threonine 23 in the vegetative events examined in this study we cannot dismiss a role for phosphorylation of the site since the isoleucine substitution may have abrogated the requirement for phosphorylation.
A cyclin-binding RRL motif is al so contained within papillomavirus E1 proteins and the tripeptide is the basis for association with multiple CDK-cyclin complexes (Ma et al., 1999). CDK phoshorylation of E1 determines a nuclear localization of the protein, where it is required to support viral DNA replication (Deng et al., 2004). The homology between this region in E1 and E1^E4, and cellular proteins that bind CDK-cyclins (e.g. p21) is high (Fig. 9), and whilst it is possible that E4 might antagonize E1 function by competition of CDK targeting our present study does not provide evidence that the role of E1 in viral DNA replication is regulated by E1^E4. Perhaps the ability of E1^E4 to associate with CDK activity is a mechanism of regulating the function of binding partners of E1^E4. Whilst an appreciation of the validity of this mechanism will require greater knowledge of E1^E4 targets in natural infections (the number of known E1^E4 targets is small (Bell, Martin, and Roberts, 2007; Doorbar et al., 2000)), it has been shown that the association between E1 and CDK-cyclin complexes enables phosphorylation of E2 when bound to E1 (Ma et al., 1999).
Figure 9. Homology between amino acid sequence of HPV E1 and E1^E4 proteins containing RXL motifs.

Comparison of the amino acids sequences of E1 and E1^E4 proteins that contain an RXL tripeptides (Cy motif) and the cellular factor p21, are shown with sequence identity, or close conservation of amino acid type, highlighted by grey-shading. Numbers indicate amino acid positions.
In conclusion, a CDK-bipartite consensus recognition motif is a common feature of E4 proteins of HPV types within the alpha genus, and thus suggests it serves an important role in E4 function in the HPV life cycle. Whilst in primary keratinocytes grown in monolayer cell culture, this motif is a key determinant in the E4 G2/M arrest function, our current study does not identify a role for this E4 motif in early vegetative events such as viral genome amplification or L1 capsid production, and future studies are necessary to establish if this conserved E4 motif has a role in the very late stages of the HPV life cycle.
MATERIALS AND METHODS
Cell culture and cell transfections
The SV40-immortalized simian kidney epithelial cell line COS-1, HeLa cervical tumour keratinocytes, the adenovirus 5 E1A-transformed epithelial cell lines 911 and J2-3T3 mouse fibroblasts were maintained in tissue culture as described previously (Knight et al., 2004).
Transient transfections of established cell lines were performed using the transfection protocol described previously (Bell, Martin, and Roberts, 2007). Routinely, cells were harvested at between 36 and 48 h post-transfection for further analysis. The isolation of normal primary human foreskin keratinocytes (HFK) from neonatal human foreskin epithelia and their subsequent maintenance in SFM keratinocyte growth media (Invitrogen, Paisley, Scotland, UK) has been described elsewhere (Ruesch and Laimins, 1998). Neonate foreskin circumcisions were collected from patients attending a G. P. practice with written informed parental consent (ethical approval number 06/Q1702/45).
Transfection of HFKs with HPV18 genomes has been described elsewhere (Wilson, Fehrmann, and Laimins, 2005). Briefly, plasmids containing the wild-type and mutant HPV18 genomes were digested with EcoRI to release the viral genome, recircularized in the presence of T4 DNA ligase and co-transfected with a plasmid encoding the neomycin resistance gene into second passage HFKs using FuGENE 6 transfection reagent (Roche, Welwyn Garden City, UK). One day after transfection, cells were seeded onto feeder layers of irradiated J2-3T3 cells and grown in the presence of G418 antibiotic (PAA Laboratories, Yeovil, UK) in foetal calf serum-containing media (E medium) for 8 days. Emerging cell colonies were pooled and expanded on irradiated J2-3T3 cells as described previously (Wilson et al., 2007).
Construction of codon-optimized HPV18 E1^E4 cDNA
A synthetic DNA sequence encoding the HPV18 E1^E4 ORF was generated using a series of oligonucleotides (18F1, 18F2, 18F3, 18R1 and 18R2; sequences are given in Table 1). These oligonucleotides were combined in a polymerase chain reaction (PCR) at a concentration of 25 pM, along with two amplifying oligonucleotides, HPV18CO-HindIIIF and HPV18CO-XHOIR (Table 1), both at 2.5 μM. The PCR product was restriction digested with HindIII and XhoI and ligated into appropriately prepared pcDNA™ 3.1/Hygro(+) plasmid (Invitrogen). Most codons were switched to that of the most frequently used codons found within the protein coding sequences of the human genome, as based on NCBI statistics (http://www.kazusa.or.jp/codon/cgibin/showcodon.cgi?species=9606).
Table 1.
Primers used for codon optimization of HPV18 E1^E4 cDNA sequence.
| Primer | Sequence (5′ to 3′) |
|---|---|
| 18F1 | ATGGCCGACCCCGAGGTGCCCGTGACAACCAGGTACCCTCTGCTCAGCCTGCTCAACAGCTACAGCACC |
| 18F2 | CCCTGGGCCCCCCAGAGGCCCACCGCCAGGAGGAGGCTGCTGCACGACCTGGACACCGTGGACAGCAGG |
| 18F3 | CACTTCAGCGTGCAGCTGCACCTGCAGGCCACCACCAAGGACGGGAACAGCGTGGTGGTGACCCTGAGGCTGTAG |
| 18R1 | GGGCCTCTGGGGGGCCCAGGGACAGGGGGCGGGGATCCTGTGGGGGGGGGTGCTGTAGCTGTTGAGCAG |
| 18R2 | GTGCAGCTGCACGCTGAAGTGGGTGCTCAGGTCCACGATGCTGCTCCTCCTGCTGTCCACGGTGTCCAG |
| 18CO-HindIIIF | GCGCAAGCTTCCACCATGGCCGACCCCGAGGTGCCC |
| 18CO XHOIR | GCGCCTCGAGCTACAGCCTCAGGGTCACCA |
Generation of HPV18 E4 mutations and construction of mutant HPV18 genomes
Nucleotide substitutions and deletions in the codon-optimized HPV18 E1^E4 coding sequence and in the E1^E4 sequence of the pGEX3T-HPV18 E1^E4 plasmid (Bell, Martin, and Roberts, 2007) were generated using the QuikChange™ site-directed mutagenesis kit (Stratagene, La Jolla, CA). The nucleotide changes made in the E4 ORF of the HPV18 genome were silent in the overlapping E2 gene, and mutagenesis was performed as described elsewhere (Wilson et al., 2007). Mutations were confirmed by bidirectional DNA sequencing, and for those mutations constructed in the HPV18 genome, the complete genomes were sequenced to also confirm their integrity outside the site of mutation.
The primer sequences used in the site-directed mutagenesis reactions are given in Table 2. The oligonucleotides used to construct mutations in the complete genomes were also used to generate the corresponding changes in the E1^E4 sequence of pGEX3T-HPV18 E1^E4, and in the plasmid vector pMC3-HPV18 E1^E4 used to generate the recombinant adenoviruses.
Table 2.
Primers used for site-directed mutagenesis of HPV18 E4 sequence.
| Primera | Sequence (5′ to 3′) |
|---|---|
| T23I | GCTACAGCATCCCACCTACAGG |
| RXL1 | CCACCGCCAGGCATAGGCGGCTGCACGACCTG |
| RXL2 | CCGCCAGGAGGCCGCTGCGGCACGACCTGGAC |
| RXLD | CCACCGCCAGGCATCCGCGGCGGCACGACCTGGAC |
| Δ21-23 | CTCAGCCTGCTCAACAGCCCACCTCACAGGATCCCC |
| S22N | GCTTGTTAAACAGCTACAACACACCCCCTCACCGT |
| ∞ T23I | GTTAAACAGCTACAGCATACCCCCTCACCG |
| ∞ RXL1 | CCTACGGCCAGACATCGGCGGCTACACGACCTG |
| ∞ RXL2 | GCCAGACGTCCGCTGCGACACGACCTG |
| ∞ RXLD | GACCTACGGCCAGACATCCGCGGCGACACGACCTGGAC |
Nucleotides underlined denotes altered nucleotide.
Primers used to construct mutations in GST-HPV18E1^E4 coding sequence, recombinant adenovirus-HPV18 E1^E4 sequence and in HPV18 genomic DNA.
Generation of recombinant adenoviruses
Recombinant adenoviruses expressing HPV18 E1^E4 wild type and mutant proteins were produced by the method of homologous recombination as described elsewhere (Roberts et al., 2003). Briefly, the HPV18 E1^E4 cDNA was sub-cloned into the BglII sites of the plasmid transfer vector pMC3 to produce pMC3-18E1^E4, and co-transfected with the plasmid containing the adenovirus type 5 E1 deleted DNA (pJM17) into 911 cells using FuGENE 6 transfection reagent. Once cytopathic effect was observed, the cells were collected by scraping and pelleted by centrifugation. Virus was released by multiple cycles of freezing and thawing and this initial virus stock was used to isolate individual plaques that were expanded to produce larger seed stocks before titration to determine particle-forming units per ml.
HFK cells grown in SFM keratinocyte growth media were infected at a multiplicity of infection of 50 for 2 h and then incubated in the presence of SFM media prior to harvesting.
Cell cycle analysis of infected cells by flow cytometry, and statistical analysis of the data, has been described by us previously (Knight et al., 2004).
Bacterial expression of glutathione-S-transferase E4 proteins
Escherichia coli BL21 cells (Stratagene) containing the various pGEX3T-E4 plasmids (Bell, Mart in, and Roberts, 2007) were grown for 16 h at 37°C in 50 ml Luria-Bertani (LB) media with 1% glucose and 50 μg/ml ampicillin. This culture was diluted into 200 ml of LB media and incubated at 37°C for 2 h, and expression induced for 3 h by addition of 50 μM isopropyl-β-D-thiogalactopyranoside (Sigma -Aldrich, Gillingham, UK) and growth at 30°C. Bacterial cells were pelleted at 4°C and resuspended in 8 ml of bacterial lysis (BL) buffer (phosphate-buffered saline (PBS), 1% Triton X-100 (vol/vol), supplemented with Complete™ protease inhibitor cocktail (Roche)). After brief sonication (2 periods of 30 s), insoluble material was removed by centrifugation and soluble GST proteins were immobilised on glutathione S-agarose beads (Sigma-Aldrich) by rotation at 4°C for 1 h. The beads were washed 5 times with BL buffer and GST proteins eluted into BL buffer containing 50 mM reduced glutathione. Soluble proteins were dialysed at 4°C in 50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 2 mM DTT and protease inhibitor cocktail.
Coprecipitation assays using recombinant GST-18 E1^E4 proteins
For coprecipitation of cyclin proteins, HeLa cells were lysed in 0.1% NP40, 50 mM HEPES pH 7.0, 250 mM NaCl, plus protease inhibitors. Cell lysates (1 mg) was mixed with 5 μg of soluble GST proteins at 4°C for 2 h. Glutathione S-agarose beads were added to each sample and GST protein complexes isolated by rotation at 4°C for 90 min. After washing the beads six times in cell lysis buffer, proteins were solubilized in Laemmli sample buffer (Bio-Rad, Hemel Hempstead, UK), boiled for 5 min and separated by electrophoresis on a 12% sodium dodecyl sulphate (SDS) polyacrylamide gel, and analysed by western blotting.
Coprecipitation assays for detection of CDK-associated activity were performed using HeLa cell lysates prepared in 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP40, and protease inhibitor cocktail. Lysates (500 μg) were mixed with 500 ng of GST fusion protein immobilized to glutathione-S-agarose beads at 4°C with rotation for 2 h. Beads were washed three times in the cell lysis buffer and twice in kinase reaction buffer containing 20 mM HEPES pH 7.4, 150 mM NaCl, 10 mM MgCl2, 1 mM DTT and protease inhibitors. In vitro phosphorylation assays were performed in the kinase reaction buffer containing 50 mM ATP, 10 μCi [γ-32P]-ATP (3000Ci/mMol, Perkin Elmer) wit h 5 μg Histone H1 (Sigma Aldrich), incubated at 30°C for 15–30 min and stopped by addition of Laemmli sample loading buffer. To examine for CDK-specific activity, in vitro phosphorylation assays were performed in the presence of the CDK inhibitor roscovitine in DMSO (Sigma -Aldrich), used at 50 μM; DMSO alone was added to control reactions.
Western blotting and immunofluorescence microscopy
Cell lysates were prepared for SDS-polyacrylamide gel electrophoresis by sonication of cells in 8M urea, 25 mM Tris-HCl pH 8.0, 0.15 M β–mercaptoethanol and protease inhibitors, or by boiling in Laemmli sample buffer. Insoluble material was removed by high-speed centrifugation and equal amounts of protein subjected to electrophoresis on SDS-polyacrylamide gels. Separated proteins were transferred to a nitrocellulose membrane, and probed with the appropriate antibody. Primary antibodies were detected with horseradish peroxidase-conjugated anti-mouse or-rabbit immunoglobulins (Sigma-Aldrich), and membranes were subjected to chemiluminescence detection (Amersham Pharmacia). HPV18 E1^E4 protein was detected using either a mouse monoclonal antibody (MAb 1D11, 1:1 dilution) (Roberts et al., 2003) or a rabbit polyclonal antibody raised against a GST-HPV18 E1^E4 fusion protein, used at a dilution of 1:500 (Wilson et al., 2007). Cyclin proteins were recognized using cyclin B1 (V152) and cyclin A (AT10.3) mouse MAbs obtained from Cancer Research UK Research Services, and a cyclin E mouse MAb (HE12, Abcam, Cambridge, UK); all used at a working dilution of 1:1000. An involucrin-specific MAb was used at 1:200 dilution (Sigma-Aldrich SY5). To control for equal loading, β-actin levels were determined by using an anti-actin MAb (1:40,000 dilution; Sigma-Aldrich).
For immunofluorescence microscopy, cells grown on glass slides, transfected with the appropriate plasmids, were fixed in 4% (wt/vol) paraformaldehyde in PBS for 5 min and per meabilized in acetone at −20°C for 10 min, or in cold methanol for 10 min. HPV18 E1^E4 was detected using the anti-GST-HPV18 E1^E4 rabbit polyclonal antibody or the 1D11 mouse MAb. Cyclin A was detected with the AT10.3 MAb or a rabbit polyclonal (sc-596, Santa Cruz Biotechnology), cyclin B1 with MAb AB3GNS1 (Neomarkers, Fremont, CA) or a rabbit polyclonal (#7957, Abcam Ltd, Cambridge UK) and cyclin E with HE12. Dual-staining for E4 and CDK1 was carried out using the 1D11 mouse MAb and a CDK1 rabbit polyclonal antibody (Ab-1, Cal Biochem). Immune complexes were recognized using the appropriate combinations of rabbit and mouse IgG specific Alexa ® 488 or 594 conjugates (Molecular probes Inc, Eugene, OR) and nuclei counter stained with 4′, 6′-diamidino-2-phenylindole (DAPI, Sigma -Aldrich). Epifluorescence microscopy was performed on a Nikon E600 microscope and images captured using a Nikon DXM1200F digital camera. Confocal microscopy was performed on a Zeiss LSM 510 META confocal microscope using multitrack imaging.
In vitro E4 phosphorylation assays and mass spectrometry
Kinase assays were carried out using recombinant active kinases p42 MAPK (5 ng, New England Biolabs (NEB), Hitchin, UK), CDK1 (10 ng, NEB) and CDK2 (0.3 ng, NEB) in the appropriate buffer containing 50 mM ATP, 10 μCi [γ-32P]-ATP (3000Ci/mMol) with 5 μg histone H1 or 5 μg of soluble GST protein as substrates. The in vitro kinase reactions were incubated at 30°C for 15–30 min and stopped by addition of Laemmli sample loading buffer. Samples were heated at 100°C for 10 min and resolved on a 12% SDS-polyacrylamide gel. The proteins were fixed and visualized by addition of fixation buffer (1% [wt/vol] Brilliant Blue (Sigma Aldrich), 40% [vol/vol] ethanol, 10% [vol/vol] acetic acid), gels dried and autoradiography performed.
For analysis of the site of phosphorylation, the kinase reaction was performed in the absence of [γ-32P]-ATP and the Coomassie-stained GST-HPV18 E4 band was extracted from the gel, digested with trypsin and analysed by mass spectrometry. Mass spectrometry was carried out by the method of Neutral Loss-Electron Capture Dissociation (NL-ECD MS/MS) by the School of Biosciences, University of Birmingham, as described elsewhere (Sweet, Creese, and Cooper, 2006). Briefly, tryptic peptides separated by liquid chromatography were introduced via a nanospray source into a Fourier transformion cyclotron resonance mass spectrometer where the mass of each peptide was calculated and ions then subjected to collision-induced disassociation. The mass spectrometry data was scanned for site-determining ions whose presence indicates phosphorylation of a particular residue (Beausoleil et al., 2006).
Differentiation of genome-containing cell lines by suspension in methylcellulose and southern analysis
Cell differentiation was induced in monolayer cultures of keratinocytes containing wild-type and mutant HPV18 genomes by first removing the J2-3T3 feeder layer, harvesting the keratinocytes and transferring them as a single cell suspension to 1.5% methylcellulose prepared in E medium as described previously (Wilson, Fehrmann, and Laimins, 2005). Cells were harvested at 24 and 48 h by centrifugation and washed extensively in PBS (Wilson et al., 2007).
For Southern analysis, total DNA was extracted from HPV18-containing keratinocytes cells grown in monolayer culture, or following suspension in methylcellulose, using a protocols described elsewhere (Wilson, Fehrmann, and Laimins, 2005; Wilson et al., 2007). DNA samples were analysed by Southern analysis using a [α32P]-dCTP-labelled HPV18 genomic probe described previously (Wilson, Fehrmann, and Laimins, 2005). To calculate copy number of HPV18 genomes, standard amounts of the HPV18 genomic DNA-containing plasmid (pGEMII-HPV18, (Wilson et al., 2007)) representing a range of copy numbers/cell were digested with EcoRI to release the complete HPV18 genome and used as copy number controls on the Southern blots. Quantification of band intensities was obtained using a Storm860 Phosphor Imager ® (GE Healthcare, Chalfont St Giles, UK) and ImageQuant 5.0 software (GE Healthcare).
Organotypic raft culture
Organotypic raft cultures were prepared as previously described (Wilson et al., 2007) and harvested after 13 days of growth. After fixation in formaldehyde, the rafts were embedded in paraffin and 4 μm sections stained with haematoxyl in and eosin to assess morphology (Propath UK Ltd). For immunohistochemistry, slides were de-paraffinized and low-temperature antigen retrieval performed as described previously (Watson et al., 2002). HPV18 E4 was detected using the rabbit polyclonal antibody, the mouse monoclonal antibody clone 5A3 (NovaCastra Laboratories) was used to detect HPV18 L1, and cyclin B recognized by a rabbit polyclonal antibody (clone H-433, Santa Cruz Biotechnology Inc.). Each cell line was grown in organotypic raft culture at least three times.
Acknowledgments
We thank Dr Joseph Spitzer and his patients for the collection and donation of foreskin tissue. We are grateful to Cleidiane Zamronio (School of Biosciences, University of Birmingham) for help with the analysis of the mass spectrometry data. We acknowledge Gordon Ryan (School of Cancer Sciences, University of Birmingham) for his technical guidance of the generation of HPV-keratinocyte cell lines. We are also grateful to staff at Propath UK Ltd (Hereford, UK) for their assistance in processing the organotypic rafts, and to Naheema Gordon and Soaud Messahel (School of Cancer Sciences, University of Birmingham) for their excellent technical support of immunohistochemistry.
This work was supported by a Cancer Research UK Programme grant to S.R. (C427/A3919) and A.G.P was supported by a Cancer Research UK Studentship (C427/A5527). L.A.L. was supported by National Institute of Health (R37CA74202).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen JL, Le Rouzic E, Planelles V. HIV-1 Vpr: mechanisms of G2 arrest and apoptosis. Exp Mol Pathol. 2008;85(1):2–10. doi: 10.1016/j.yexmp.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24(10):1285–92. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Bell I, Martin A, Roberts S. The E1^E4 protein of human papillomavirus interacts with the serine-arginine-specific protein kinase SRPK1. J Virol. 2007;81(11):5437–48. doi: 10.1128/JVI.02609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995;87(11):796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V. A review of human carcinogens--Part B: biological agents. Lancet Oncol. 2009;10(4):321–2. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- Breitburd F, Croissant O, Orth G. Expression of human papillomavirus type-1 E4 gene products in wart s. In: Steinberg BM, Brandsma J, Taichman LB, editors. Papillomaviruses. Vol. 5. Cold Spring Harbor press; New York: 1987. pp. 115–122. [Google Scholar]
- Bryan JT, Han A, Fife KH, Brown DR. The human papillomavirus type 11 E1E4 protein is phosphorylated in genital epithelium. Virology. 2000;268(2):430–9. doi: 10.1006/viro.1999.0173. [DOI] [PubMed] [Google Scholar]
- Chow LT, Duffy AA, Wang HK, Broker TR. A highly efficient system to produce infectious human papillomavirus: Elucidation of natural virus-host interactions. Cell Cycle. 2009;8(9):1319–23. doi: 10.4161/cc.8.9.8242. [DOI] [PubMed] [Google Scholar]
- Chulu JL, Huang WR, Wang L, Shih WL, Liu HJ. Avian reovirus nonstructural protein p17-induced G(2)/M cell cycle arrest and host cellular protein translation shutoff involve activation of p53-dependent pathways. J Virol. 2010;84(15):7683–94. doi: 10.1128/JVI.02604-09. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Davy C, Doorbar J. G2/M cell cycle arrest in the life cycle of viruses. Virology. 2007;368(2):219–26. doi: 10.1016/j.virol.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy CE, Ayub M, Jackson DJ, Das P, McIntosh P, Doorbar J. HPV16 E1^E4 protein is phosphorylated by Cdk2/cyclin A and relocalizes this complex to the cytoplasm. Virology. 2006;349(1):230–44. doi: 10.1016/j.virol.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Davy CE, Jackson DJ, Raj K, Peh WL, Southern SA, Das P, Sorathia R, Laskey P, Middleton K, Nakahara T, Wang Q, Masterson PJ, Lambert PF, Cuthill S, Millar JB, Doorbar J. Human papillomavirus type 16 E1^E4-induced G2 arrest is associated with cytoplasmic retention of active Cdk1/cyclin B1 complexes. J Virol. 2005;79(7):3998–4011. doi: 10.1128/JVI.79.7.3998-4011.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy CE, Jackson DJ, Wang Q, Raj K, Masterson PJ, Fenner NF, Southern S, Cuthill S, Millar JB, Doorbar J. Identification of a G(2) arrest domain in the E1^E4 protein of human papillomavirus type 16. J Virol. 2002;76(19):9806–18. doi: 10.1128/JVI.76.19.9806-9818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin BY, Jin G, Wheeler CG, Ma T, Harper JW, Broker TR, Chow LT. Cyclin/CDK regulates the nucleocytoplasmic localization of the human papillomavirus E1 DNA helicase. J Virol. 2004;78(24):13954–65. doi: 10.1128/JVI.78.24.13954-13965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Doorbar J, Li B, Zhou F, Gu W, Zhao L, Saunders NA, Frazer IH, Zhao KN. Expression of papillomavirus L1 proteins regulated by authentic gene codon usage is favoured in G2/M-like cells in differentiating keratinocytes. Virology. 2010;399(1):46–58. doi: 10.1016/j.virol.2009.12.021. [DOI] [PubMed] [Google Scholar]
- Doorbar J, Campbell D, Grand RJ, Gallimore PH. Identification of the human papilloma virus-1a E4 gene products. Embo J. 1986;5(2):355–62. doi: 10.1002/j.1460-2075.1986.tb04219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorbar J, Elston RC, Napthine S, Raj K, Medcalf E, Jackson D, Coleman N, Griffin HM, Masterson P, Stacey S, Mengistu Y, Dunlop J. The E1E4 protein of human papillomavirus type 16 associates with a putative RNA helicase through sequences in its Cterminus. J Virol. 2000;74(21):10081–95. doi: 10.1128/jvi.74.21.10081-10095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorbar J, Ely S, Sterling J, McLean C, Crawford L. Specific interaction between HPV-16 E1-E4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature. 1991;352(6338):824–7. doi: 10.1038/352824a0. [DOI] [PubMed] [Google Scholar]
- Fang L, Budgeon LR, Doorbar J, Briggs ER, Howett MK. The human papillomavirus type 11 E1/\E4 protein is not essential for viral genome amplification. Virology. 2006;351(2):271–9. doi: 10.1016/j.virol.2006.01.051. [DOI] [PubMed] [Google Scholar]
- Hebner CM, Laimins LA. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev Med Virol. 2006;16(2):83–97. doi: 10.1002/rmv.488. [DOI] [PubMed] [Google Scholar]
- Hindmarsh PL, Laimins LA. Mechanisms regulating expression of the HPV 31 L1 and L2 capsid proteins and pseudovirion entry. Virol J. 2007;4:19. doi: 10.1186/1743-422X-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight GL, Grainger JR, Gallimore PH, Roberts S. Cooperation between different forms of the human papillomavirus type 1 E4 protein to block cell cycle progression and cellular DNA synthesis. J Virol. 2004;78(24):13920–33. doi: 10.1128/JVI.78.24.13920-13933.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight GL, Turnell AS, Roberts S. Role for Wee1 in inhibition of G2-to-M transition through the cooperation of distinct human papillomavirus type 1 E4 proteins. J Virol. 2006;80(15):7416–26. doi: 10.1128/JVI.00196-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Zou N, Lin BY, Chow LT, Harper JW. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc Natl Acad Sci U S A. 1999;96(2):382–7. doi: 10.1073/pnas.96.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara T, Nishimura A, Tanaka M, Ueno T, Ishimoto A, Sakai H. Modulation of the cell division cycle by human papillomavirus type 18 E4. J Virol. 2002;76(21):10914–20. doi: 10.1128/JVI.76.21.10914-10920.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara T, Peh WL, Doorbar J, Lee D, Lambert PF. Human papillomavirus type 16 E1^E4 contributes to multiple facets of the papillomavirus life cycle. J Virol. 2005;79(20):13150–65. doi: 10.1128/JVI.79.20.13150-13165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasseri M, Hirochika R, Broker TR, Chow LT. Ahuman papilloma virus type 11 transcript encoding an E1--E4 protein. Virology. 1987;159(2):433–9. doi: 10.1016/0042-6822(87)90482-x. [DOI] [PubMed] [Google Scholar]
- Peh WL, Brandsma JL, Christensen ND, Cladel NM, Wu X, Doorbar J. The viral E4 protein is required for the completion of the cottontail rabbit papillomavirus productive cycle in vivo. J Virol. 2004;78(4):2142–51. doi: 10.1128/JVI.78.4.2142-2151.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peh WL, Middleton K, Christensen N, Nicholls P, Egawa K, Sotlar K, Brandsma J, Percival A, Lewis J, Liu WJ, Doorbar J. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J Virol. 2002;76(20):10401–16. doi: 10.1128/JVI.76.20.10401-10416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj K, Berguerand S, Southern S, Doorbar J, Beard P. E1 empty set E4 protein of human papillomavirus type 16 associates with mitochondria. J Virol. 2004;78(13):7199–207. doi: 10.1128/JVI.78.13.7199-7207.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S, Ashmole I, Johnson GD, Kreider JW, Gallimore PH. Cutaneous and mucosal human papillomavirus E4 proteins form intermediate filament-like structures in epithelial cells. Virology. 1993;197(1):176–87. doi: 10.1006/viro.1993.1578. [DOI] [PubMed] [Google Scholar]
- Roberts S, Ashmole I, Rookes SM, Gallimore PH. Mutational analysis of the human papillomavirus type 16 E1^E4 protein shows that the C terminus is dispensable for keratin cytoskeleton association but is involved in inducing disruption of the keratin filaments. J Virol. 1997;71(5):3554–62. doi: 10.1128/jvi.71.5.3554-3562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S, Hillman ML, Knight GL, Gallimore PH. The ND10 component promyelocytic leukemia protein relocates to human papillomavirus type 1 E4 intranuclear inclusion bodies in cultured keratinocytes and in warts. J Virol. 2003;77(1):673–84. doi: 10.1128/JVI.77.1.673-684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S, Kingsbury SR, Stoeber K, Knight GL, Gallimore PH, Williams GH. Identification of an arginine-rich motif in human papillomavirus type 1 E1;E4 protein necessary for E4-mediated inhibition of cellular DNA synthesis in vitro and in cell s. J Virol. 2008;82(18):9056–64. doi: 10.1128/JVI.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruesch MN, Laimins LA. Human papillomavirus oncoproteins alter differentiation-dependent cell cycle exit on suspension in semisolid medium. Virology. 1998;250(1):19–29. doi: 10.1006/viro.1998.9359. [DOI] [PubMed] [Google Scholar]
- Ruesch MN, Stubenrauch F, Laimins LA. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J Virol. 1998;72(6):5016–24. doi: 10.1128/jvi.72.6.5016-5024.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet SM, Creese AJ, Cooper HJ. Strategy for the identification of sites of phosphorylation in proteins: neutral loss triggered electron capture dissociation. Anal Chem. 2006;78(21):7563–9. doi: 10.1021/ac061331i. [DOI] [PubMed] [Google Scholar]
- Takeda DY, Wohlschlegel JA, Dutta A. A bipartite substrate recognition motif for cyclin-dependent kinases. J Biol Chem. 2001;276(3):1993–7. doi: 10.1074/jbc.M005719200. [DOI] [PubMed] [Google Scholar]
- Toyoshima-Morimoto F, Taniguchi E, Shinya N, Iwamatsu A, Nishida E. Polo-like kinase 1 phosphorylates cyclin B1 and targets it to the nucleus during prophase. Nature. 2001;410(6825):215–20. doi: 10.1038/35065617. [DOI] [PubMed] [Google Scholar]
- Wang HK, Duffy AA, Broker TR, Chow LT. Robust production and passaging of infectious HPV in squamous epithelium of primary human keratinocytes. Genes Dev. 2009a;23(2):181–94. doi: 10.1101/gad.1735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Kennedy A, Das P, McIntosh PB, Howell SA, Isaacson ER, Hinz SA, Davy C, Doorbar J. Phosphorylation of the human papillomavirus type 16 E1--E4 protein at T57 by ERK triggers a structural change that enhances keratin binding and protein stability. J Virol. 2009b;83(8):3668–83. doi: 10.1128/JVI.02063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RA, Rollason TP, Reynolds GM, Murray PG, Banks L, Roberts S. Changes in expression of the human homologue of the Drosophila discs large tumour suppressor protein in high-grade premalignant cervical neoplasias. Carcinogenesis. 2002;23(11):1791–6. doi: 10.1093/carcin/23.11.1791. [DOI] [PubMed] [Google Scholar]
- Wilson R, Fehrmann F, Laimins LA. Role of the E1--E4 protein in the differentiation-dependent life cycle of human papillomavirus type 31. J Virol. 2005;79(11):6732–40. doi: 10.1128/JVI.79.11.6732-6740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Ryan GB, Knight GL, Laimins LA, Roberts S. The full-length E1E4 protein of human papillomavirus type 18 modulates differentiation-dependent viral DNA amplification and late gene expression. Virology. 2007;362(2):453–60. doi: 10.1016/j.virol.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Takeda DY, Dutta A. Mutational analysis of the Cy motif from p21 reveals sequence degeneracy and specificity for different cyclin-dependent kinases. Mol Cell Biol. 2001;21(15):4868–74. doi: 10.1128/MCB.21.15.4868-4874.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu WJ, Peng SW, Sun XY, Frazer I. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. J Virol. 1999;73(6):4972–82. doi: 10.1128/jvi.73.6.4972-4982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]