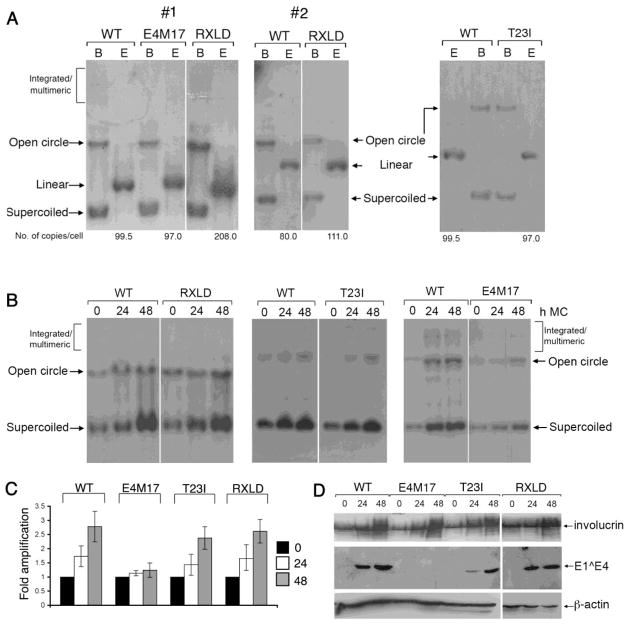
Figure 7. The CDK-bipartite recognition motif in E4 is not essential for establishment of the viral genome or differentiation-dependent viral genome amplification.
(A) Southern blot analysis of HFK stably transfected with HPV18 wild-type (WT), RXLD and E4M17 mutant genomes. Equal amounts of total DNA was digested with DpnI to remove residual input bacterial DNA and either BglII (B), which does not have a site in the HPV18 genome or EcoR1 (E), which has a single recognition site in the HPV18 genome and produces linear forms of the viral genome. HPV18 episomes migrate as supercoils and open circles. Whilst in one donor (#1), the RXLD genomes established at a higher copy number than the wild type genomes this was not a consistent observation across all donors (e.g. 2). (B) Southern analysis of equal amounts of total genomic DNA isolated from keratinocyte cell lines containing wild-type (WT) or mutant (T23I, RXLD, E4M17) HPV18 genomes grown in monolayer cell culture (0 h) or suspended in methylcellulose (MC) for 24 and 48 h. DNA was digested with DpnI and BglII and HPV18 episomes migrate as supercoils and open circles. (C) Bargraph of phosphoimager analysis of Southern blots to show levels of HPV18 DNA genome amplification. Data were derived from at least fifteen (WT and RXLD) or six (E4M17 and T23I) experiments of each cell line in a total of three different donor keratinocytes. (D) Western blot analysis of the total protein extracts isolated from the different HPV18 genome-containing cell lines to determine expression levels of HPV18 E1^E4 protein. The induction of differentiation was monitored by determination of the expression levels of the epidermal differentiation-marker involucrin. Levels of β-actin were used as a loading control.