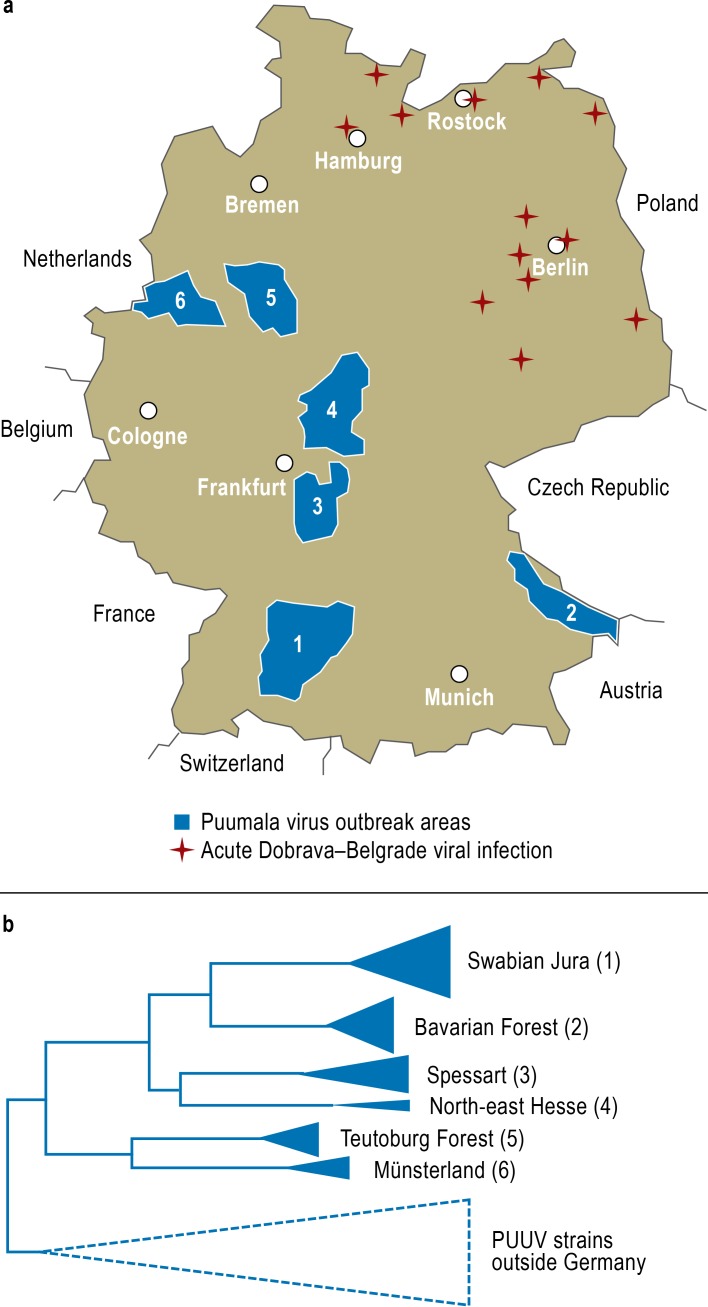
Figure 2.
Molecular epidemiological cadastre of hantavirus strains in Germany
a) Areas of Puumala virus outbreaks in 2010 (blue). In these areas, viral nucleic acids from both bank voles and patients were analyzed and compared. Places with evidence of patients with Dobrava–Belgrade viral infection in the north and east of Germany are also shown (from: [18]: Ettinger J, Hofmann J, Enders M, et al.: Multiple synchronous outbreaks of Puumala virus, Germany, 2010. Emerg Infect Dis 2012; 18: 1461–4; with the addition of new, unpublished data in the possession of the present authors).
b) Schematic representation of the phylogenetic relationships between the viral strains from six Puumala virus outbreak areas, from which viral sequences from patients and local bank voles were analyzed. The viral strains of each outbreak area form their own “molecular cluster,” which is different from that of the neighboring outbreak areas (from [34]: Epidemiologisches Bulletin des Robert Koch-Institutes: Molekulare Unterscheidbarkeit der zirkulierenden Hantavirus-Stämme in den verschiedenen Ausbruchsregionen Deutschlands. Epidem Bull 2012, Nr. 25, 228–31; reproduced by kind permission of the Robert Koch Institute)