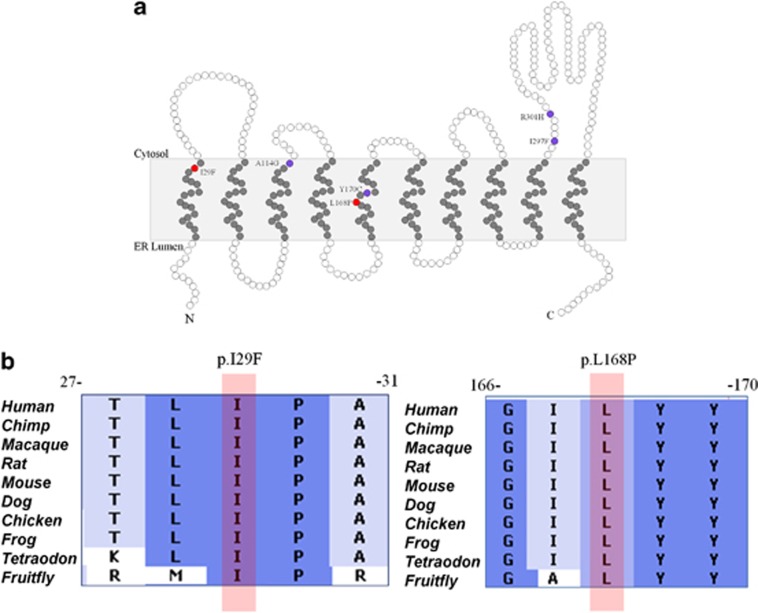
Figure 3.
Schematic representation of DPAGT1 protein and location of the mutations. (a) The white circles indicate hydrophilic residues, whereas gray circles indicate the hydrophobic residues of the protein. The mutations found in this study are indicated by red circles (p.I29F and p.L168P), located in the first and fifth transmembrane domains of the protein, respectively. The mutations that are already described in literature are shown by purple circles. (b) Positions of the amino-acid substitutions I29F and L168P (indicated by pink bar), which are highly conserved down to the Fruitfly, predicting the disruptive effect on the normal function of the protein.