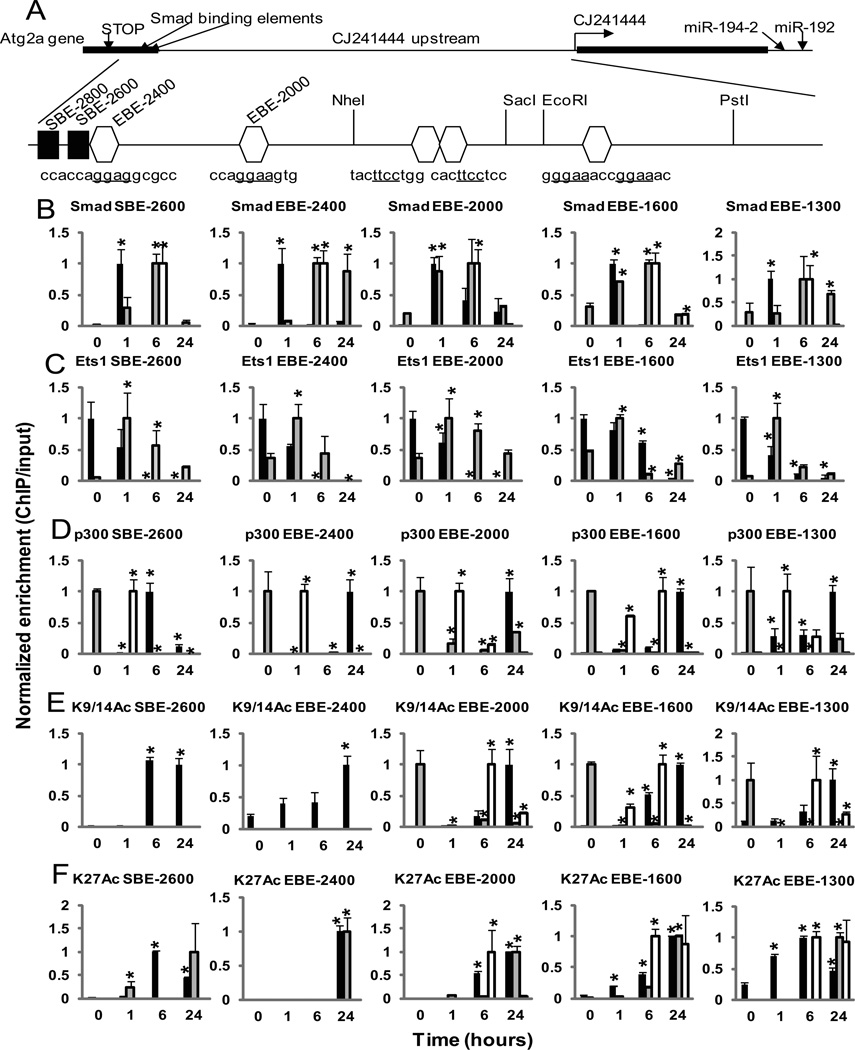
Fig. 4. Chromatin Immunoprecipitation (ChIP) assays reveal TGF-β induced changes in the occupancies of Smads and Ets-1 and histone acetylation in the upstream region of miR-192.
(A) Schematic representation of the upstream region of miR-192. A transcript (CJ241444, CJ24) as well as the Atg2a gene are located in the upstream region of the miR-192 gene. Smad binding elements (SBE-2800 and SBE-2600) are found in the 3’UTR of Atg2a. Four clusters of potential Ets-1 binding sites (EBE-2400, EBE-2000, EBE-1600, and EBE-1300) are present in the upstream region of CJ24. (B to F) Cross-linked chromatin samples from MCs treated with TGF-β for 0 to 24 hours were immunoprecipitated with indicated antibodies: Smad2 and Smad3 (Smad) (B), Ets-1 (Ets1) (C), p300 (D), acetylated histone H3K9 and H3K14 (K9/14Ac) (E), and H3K27Ac (K27Ac) (F). ChIP enriched DNA samples were amplified by qPCR using primers spanning each of these sites. Black bars, MCs treated with TGF-β alone; Gray bars, MCs treated with TGF-β and MK-2206; White bars, Ets-1-deficient MCs treated with TGF-β. Data are means ± SEM from three experiments. *P < 0.05 compared to the 0 time point by one-way ANOVA followed by Tukey’s post-hoc test analysis.