Abstract
Mycobacterium tuberculosis (Mtb) isolates with identical genotypes, found in different patients, are most likely the result of recent transmission. Mtb strains with closely related genotypes, called clonal complexes, are most likely derived from one another. We examined Mtb genotypes from southern California TB patients from 2005 through 2008 to complete the first comprehensive molecular epidemiology analysis of this complicated and ethnically diverse region. Mtb genotypes were characterized with spoligo-type and MIRU-12 typing. MIRU-VNTRplus was utilized to assign genotypes to global lineages and complete cluster analyses. Associations between patient characteristics and genotype clustering and clonal complexes were evaluated using logistic regression and frequency analysis. Of 832 Mtb isolates analyzed, 480 (58%) fell into 94 strain clusters. The majority of isolates were identified as being in the EA1 (31%), LAM (17%) and Haarlem (15%) lineages, but 13 different lineages were found in this region. TB patients with clustered isolates were more likely to be homeless (AOR 3.44, 95% CI 1.65, 7.18) and male (AOR 1.57, 95% CI 1.17, 2.10). Of the 480 clustered strains, 388 aggregated into six clonal complexes.
Over 45% of reported TB cases were clustered and likely resulted from recent transmission events. Patients with clustered Mtb isolates that were grouped into clonal complexes had unique socio-demographic characteristics. These data suggest that TB is being transmitted in relatively insular community networks defined by race/ethnicity and country of origin. The addition of clonal complex analysis to simple cluster analysis provides important public health insights into the local transmission of TB in ethnically diverse regions with diverse Mtb genotypes.
Keywords: Tuberculosis, Molecular epidemiology, Social networks, Genotypes, Spoligotype, MIRU
1. Introduction
Molecular epidemiology has revolutionized the study of tuberculosis (TB) transmission and the management of TB disease (Allix-Beguec et al., 2008; Bezanahary et al., 2008; David, 2008; Fica et al., 2008; Mathema et al., 2008). The fundamental principal behind the molecular epidemiology of TB is that, unlike other bacteria, Mtb replicates clonally and almost never transfers genes horizontally. This together with the fact that Mtb underwent a genetic bottleneck recently, makes it relatively monomorphic (Gibson et al., 2008). Detectable genetic variation between different Mtb bacilli does evolve over time, however, which means Mtb strains with identical genotypes found in different patients are most likely the result of recent transmission (Allix-Beguec et al., 2008; Bezanahary et al., 2008; David, 2008; Fica et al., 2008; Mathema et al., 2008). Furthermore, Mtb strains with genotypes shown to be closely related by phylogenetic analysis (defined as differing by only one MIRU locus in this study) were probably derived from one another, giving rise to the notion of a clonal complex of Mtb strains that may, but not necessarily do, share a recent transmission event as well (Allix-Beguec et al., 2008; Weniger et al., 2010).
From a TB management perspective, characterizing Mtb strains at the genomic level enables TB programs to track the transmission of specific Mtb strains, follow epidemics, and identify new outbreaks. Evaluating molecular epidemiology from a population genetics perspective also enables public health researchers to determine which patient factors might be associated with clusters of related strains and how interventions might be formulated to prevent those strains from being transmitted in the future (Behr and Mostowy, 2007; Garzelli and Rindi, 2012).
In the US, the national genotyping program determines Mtb genotypes primarily on the basis of spacer oligonucleotide typing (spoligotyping) (Kamerbeek et al., 1997) and mycobacterial interspersed repetitive unit (MIRU) typing (Mazars et al., 2001). In this study, we examined Mtb spoligotype and MIRU genotypes from San Diego County from 2005 through 2008, in order to complete the first comprehensive molecular epidemiology analysis of TB of this region.
San Diego County is unique in the US. Over 70% of TB cases in San Diego County occur in foreign-born patients of multiple nationalities, and TB incidence is almost double the US national average (San Diego Health and Human Service Agency (HHSA), 2011). In order to better understand the factors associated with transmission of TB in this complicated and ethnically diverse region, we completed, (i) an analysis of Mtb strain lineages to determine the diversity of strains present; (ii) an analysis of clustered vs. non-clustered strains to determine the proportion of recent transmission vs. reactivated TB cases (Small et al., 1994); and (iii) an analysis of clonal complexes within the clustered cases to determine how the recent transmission cases might be related to one another genetically and epidemiologically (Allix-Beguec et al., 2008; Weniger et al., 2010).
2. Materials and methods
2.1. Study population
San Diego is the second largest county in California with a diverse population of over three million people (United States Census Bureau, 2011) and one of the highest TB case rates in the US (6.0/ 100,000 population) (Centers for Disease Control and Prevention (CDC), 2011). Located in the southernmost portion of the state, it shares one of the world’s busiest land border-crossings with Mexico (Lange et al., 1999), and has a high rate of immigration from countries such as Vietnam and Philippines, where TB is endemic (International Community Foundation (ICF), 2010). California law (Health and Safety Code Title 17 §2505) requires that all verified cases of TB be documented and reported to the US National TB Control Program. Approximately 200–300 incident TB cases are reported in San Diego County annually (San Diego Health and Human Service Agency (HHSA), 2011).
2.2. Data source
Socio-demographic and clinical data, as well as pathogen genotypes for TB patients, included in this study were obtained from the San Diego Report of a Verified Case of Tuberculosis (RVCT) database supplemented with locally collected variables. RVCT data were collected based on US national TB surveillance guidelines and all TB cases evaluated in the study met the national surveillance TB case definition (laboratory or clinical evidence of disease caused by Mycobacterium tuberculosis complex) (Centers for Disease Control and Prevention (CDC), 1997). Pathogen genotypes and TB surveillance variables from all culture-positive cases of TB reported to the San Diego County TB program from 2005 through 2008 were included in this study. The study protocol was approved by an Institutional Review Board at the University of California, San Diego.
2.3. TB culture and species identification
Since the early 1990s, the San Diego County Public Health Laboratory obtained an Mtb isolate from over 80% of all TB patients for pathogen species identification and drug susceptibility testing (DST) (Rodwell et al., 2008). All TB isolates from patient specimens were initially identified as Mtb complex on the basis of the AccuP-robe hybridization protection assay (GenProbe, San Diego, CA, USA). Due to the fact that TB from Mycobacterium bovis is also prevalent in Southern California (Rodwell et al., 2010b; Rodwell et al., 2008), cultured isolates were further identified as either Mtb or M. bovis on the basis of culture morphologic findings, the results of the niacin strip test, the nitrate reduction test (Grange et al., 1996) or their spoligotype (Streicher et al., 2007). TB cases identified as M. bovis strains were excluded from further analysis.
2.4. Strain genotyping
Mtb genotypes were characterized using spoligotyping (Kamerbeek et al., 1997) and MIRU-12 typing (Mazars et al., 2001). Spoligotyping and MIRU were performed and reported by the Microbial Diseases Laboratory at the California Department of Public Health according to Centers for Disease Control and Prevention (CDC) guidelines (Centers for Disease Control and Prevention (CDC), 2004). Spoligotyping was completed with Luminex-based methods which detect 43 specific spacer sequences in the direct repeat locus (Cowan et al., 2004). Each spoligotype was converted from the 43-digit binary sequence to the octal format for analysis (Dale et al., 2001). MIRU-12 was completed using procedures described by Cowan et al. (2002) to determine the number of repeat sequences at the 12 MIRU loci: 02, 04, 10, 16, 20, 23, 24, 26, 27, 31, 39, and 40 (Mazars et al., 2001). We acknowledge that MIRU-15 or MIRU-24 typing has recently been adopted by most nations (including the US) to replace MIRU-12 genotyping as MIRU-12 typing plus spoligotyping has been shown to overestimate isolate clustering due to insufficient variation at the MIRU-12 loci (Supply et al., 2006). As this analysis spans the years 2005 through 2008, MIRU-24 data was not available. We discuss the potential effect of MIRU-12 genotyping on our estimates in the discussion section.
2.5. Determination of lineage and lineage evaluation
Genotype information was uploaded to the MIRU-VNTRplus web application (http://www.miru-vntrplus.org) for lineage assignment and cluster analysis (Allix-Beguec et al., 2008; Weniger et al., 2010). A strain lineage was identified for each isolate using the “similarity search” module, which compared the combined spoligotype and MIRU genotypes against a collection of 186 reference strains representing major global Mtb complex lineages (Allix-Beguec et al., 2008). A 4-step progressive approach was employed for phylogenetic lineage assignment. First, the lineage matches were based on categorical distance measures using MIRU and spoligotype sequences. In the “tree-based identification” module set to the neighbor-joining (NJ) algorithm (Saitou and Nei, 1987), any isolates still missing lineages were then analyzed for categorical distance matches to the nearest subtree using MIRU sequence alone, then MIRU and spoligotype sequences for further remaining isolates, and finally, spoligotype sequences alone for the last unassigned isolates. Characterized lineages were matched with the global repository of TB lineages (spolDB4) and strains with no match in spolDB4 were considered “orphan” strains (Brudey et al., 2006).
2.6. Cluster definition
TB cases were defined as “clustered” when ⩾2 Mtb isolates from different patients evaluated during the study period had identical spoligotype and MIRU-12 genotypes. Strains not found to have matching genotypes with any other isolates in the study period were classified as “singletons.” Clustered cases were assumed to have resulted from recently acquired, person-to-person infections while singleton cases were assumed to most likely have originated from reactivated latent tuberculosis infections acquired distantly in time and space (Mathema et al., 2008; Small et al., 1994). Patients with the earliest report date within a cluster were assumed to be the index cases, making the number of assumed transmission events in each cluster N−1 (where N is the cluster size) (Metcalfe et al., 2010; Small et al., 1994).
2.7. Characterization of clonal complexes
The genetic relatedness of strains from clustered cases, was further analyzed by calculating the frequency of locus variants using the “minimum spanning tree” (MST) module of the online bioinformatics tool, MIRU-VNTRplus. The MST analysis organized the Mtb strains of related MIRU genotypes, into “clonal complexes” as described in detail in previous studies (Allix-Beguec et al., 2008; Weniger et al., 2010). A clonal complex in this analysis was defined as a group of strains with no more than a single locus variation (SLV) (i.e., maximum threshold of one MIRU locus variation from a central strain or any another strain in the complex).
2.8. Statistical analyses
We examined the associations between clustered TB cases, and potential socio-demographic and clinical risk factors with logistic regression modeling. The outcome was clustered vs. singleton (reference) cases. Univariate logistic regression was used to describe the associations between the outcome variable and each independent variable. All variables that were statistically significant (p < 0.05) in the univariate analyses were considered for inclusion in multivariate analysis. The final model was derived by removing each variable with the highest p-value in turn until only those variables with p-values <0.05 remained.
We also examined the associations between potential socio-demographic and clinical risk factors and clonal complexes using frequency analysis (Stokes et al., 2000). Clustered cases that could not be grouped into a clonal complex (i.e., they had more than one MIRU loci different from other clonal complexes) were excluded. Frequencies of categorical variables were compared by Chi-square test, unless there were too few expected values in one or more cells, in which case Fisher’s exact test was used (Stokes et al., 2000). If one of the cells contained zero strains, then no statistical test could applied for that variable. Continuous variables were compared by the Kruskal–Wallis one-way analysis of variance (Zar, 1999).
3. Results
From 2005 through 2008, 1164 incident cases of TB were reported in San Diego County. Eighty four percent (977/1164) of the cases had isolates that were cultured successfully and were genotyped with spoligotype and MIRU typing. Of the 977 isolates genotyped, 102 were identified as M.bovis and were excluded from further analysis. A further 43 isolates were excluded because their MIRU or spoligotype data were incomplete or missing; leaving 832 patients and their Mtb isolates with completed surveillance and genotyping data in the analysis. Median age of the 832 TB patients included in the analysis was 47 years (IRQ 29,64) and 35% were female.
Of the 832 Mtb isolates examined, 480 (58%) matched the genotype of at least one other isolate, and were collated into 94 clusters. Cluster sizes ranged from 2 to 93 isolates. The remaining 352 isolates were singletons that did not match any of the other strains in the sample. The proportion of clustered TB cases did not differ substantially by year (Table 1). After accounting for one index case in each cluster, we estimated that 386/832 (46%) of reported cases likely resulted from recent transmission events.
Table 1.
Number of Mycobacterium tuberculosis isolates in San Diego County with singleton and clustered genotypesa by year of diagnosis.
| Year | All cases (% all years) |
Singleton cases (% patients) |
Clustered cases (% patients) |
|---|---|---|---|
| 2005–2008 | 832 (100) | 352 (42.3) | 480 (57.7) |
| 2005 | 203 (23.4) | 93 (45.8) | 110 (54.2) |
| 2006 | 215 (25.8) | 102 (47.4) | 113 (52.6) |
| 2007 | 216 (26.0) | 75 (34.7) | 141 (65.3) |
| 2008 | 198 (23.8) | 82 (41.4) | 116 (58.6) |
Genotypes defined spoligotype and mycobacterial interspersed repetitive units (MIRU) typing.
3.1. Lineage analysis
The spoligotypes of most of the isolates examined matched those previously described and reported in the SpolDB4 global spoligotype database. Only 91/832 (11%) isolates did not have a shared spoligotype from SpolDB4 (and were therefore designated “orphans”). Most of the orphan genotypes (N = 74) were from the 352 singleton strains. Among the 94 clustered strains, only 8 strain types were defined as orphans; representing 17/480 (4%) of the total clustered isolates.
The majority of isolates were identified as belonging to the EA1 (31%), LAM (17%) and Haarlem (15%) lineages, but overall 13 different lineages were found (Table 2). It was notable that most of the clustered and singleton isolates were found in similar proportions among the top three most prevalent lineages. Beijing strains, however, were twice as common among clustered cases compared to singleton cases and the X strains were more than twice as frequent among singleton cases compared to clustered cases (see Supplemental Table 1 for complete dataset).
Table 2.
Number of Mycobacterium tuberculosis isolates in San Diego County from 2005 through 2008, with singleton and clustered genotypesb, stratified by strain lineage.
|
Mtb lineage |
All cases N (%) |
Singleton cases N (%) |
Clustered Cases N (%) |
X2 | p |
|---|---|---|---|---|---|
| Total | 832 (100.0) |
352 (100.0) | 480 (100.0) | ||
| EAI | 257 (30.9) | 97 (27.6) | 160 (33.3) | 1.51 | 0.2199 |
| LAM | 141 (16.9) | 73 (20.7) | 68 (14.2) | 4.01 | 0.0451 |
| Haarlem | 126 (15.1) | 58 (16.5) | 68 (14.2) | 0.48 | 0.4897 |
| Beijing | 79 (9.5) | 21 (6.0) | 58 (12.1) | 6.74 | 0.0094 |
| S | 58 (7.0) | 13 (3.7) | 45 (9.4) | 8.07 | 0.0045 |
| UgandaI | 55 (6.6) | 18 (5.1) | 37 (7.7) | 1.57 | 0.2097 |
| X | 55 (6.6) | 37 (10.5) | 18 (3.8) | 12.06 | 0.0005 |
| Dehli/CAS | 28 (3.4) | 19 (5.4) | 9 (1.9) | 6.20 | 0.0127 |
| TUR | 10 (1.2) | 2 (0.6) | 8 (1.7) | 0.2054a | |
| UgandaII | 8 (1.0) | 3 (0.9) | 5 (1.0) | 1.000a | |
| Cameroon | 6 (0.7) | 6 (1.7) | 0 (0.0) | n/a | n/a |
| Ghana | 5 (0.6) | 3 (0.9) | 2 (0.4) | 0.6555a | |
| NEW-1 | 4 (0.5) | 2 (0.6) | 2 (0.4) | 1.000a |
n/a = no statistical test done due to zero value cells in row.
Fisher exact test used.
Genotypes were defined by spoligotype and mycobacterial interspersed repetitive units (MIRU) typing.
3.2. Variables associated with clustered vs. singleton tb cases
In our evaluation of socio-demographic and clinical variables associated with clustered vs. singleton TB cases, univariate analyses demonstrated that many variables were significantly associated with clustering (Table 3). After removing variables that were not significant in the multivariate logistic regression model, however, the final model included only four significant variables. Table 3 indicates that TB patients with clustered isolates were more likely to be homeless (AOR 3.44, 95% CI 1.65, 7.18), male (AOR 1.57, 95% CI 1.17, 2.10), and less likely to be older (AOR 0.90, 95% CI 0.84, 0.96). TB patients born in the Philippines were also twice as likely to have clustered Mtb isolates compared to patients born in the US (AOR 1.86, 95% CI 1.17, 2.95).
Table 3.
Univariate and multivariate logistic regression analysis of socio-demographic and clinical factors associated with Mycobacterium tuberculosis genotype clustering among 832a tuberculosis cases diagnosed in San Diego County, 2005–2008.
| Variable | N | Univariate logistic regression |
Multivariate logistic regression |
||||
|---|---|---|---|---|---|---|---|
| OR | 95% CL | p | AOR | 95% CL | p | ||
| Gender | |||||||
| Female | 292 | 1.00 | 1.00 | ||||
| Male | 540 | 1.66 | 1.24, 2.21 | 0.0006 | 1.63 | 1.20, 2.22 | 0.0020 |
| Age (per 10 years) | 832 | 0.90 | 0.84, 0.96 | 0.0025 | 0.86 | 0.80, 0.92 | <0.0001 |
| Race/ethnicity | |||||||
| White, non-Hispanic | 76 | 1.00 | |||||
| Asian | 311 | 1.22 | 0.74, 2.03 | 0.4396 | |||
| Black | 67 | 0.66 | 0.34, 1.27 | 0.2116 | |||
| Hispanic | 375 | 1.14 | 0.69, 1.87 | 0.6137 | |||
| Country of birth | |||||||
| US-born | 185 | 1.00 | |||||
| Foreign-born | 646 | 0.68 | 0.49, 0.96 | 0.0270 | |||
| Country of origin | |||||||
| U.S. | 185 | 1.00 | 1.00 | ||||
| Mexico | 258 | 0.70 | 0.47, 1.03 | 0.0671 | 0.78 | 0.52, 1.17 | 0.2270 |
| Philippines | 197 | 1.21 | 0.79, 1.85 | 0.3868 | 1.86 | 1.17, 2.95 | 0.0085 |
| Vietnam | 51 | 0.45 | 0.24, 0.83 | 0.0116 | 0.57 | 0.30, 1.09 | 0.0894 |
| Other | 141 | 0.36 | 0.23, 0.56 | <0.0001 | 0.45 | 0.28, 0.71 | 0.0008 |
| Injection drug use | |||||||
| No | 811 | 1.00 | |||||
| Yes | 21 | 1.48 | 0.59, 3.71 | 0.4020 | |||
| Non-inject drug use | |||||||
| No | 739 | 1.00 | |||||
| Yes | 93 | 1.81 | 1.14, 2.89 | 0.0125 | |||
| Excessive alcohol use | |||||||
| No | 710 | 1.00 | |||||
| Yes | 122 | 1.61 | 1.07, 2.41 | 0.0220 | |||
| Homeless | |||||||
| No | 772 | 1.00 | 1.00 | ||||
| Yes | 53 | 3.83 | 1.84, 7.95 | 0.0003 | 3.44 | 1.65, 7.18 | 0.0010 |
| Correction facility | |||||||
| No | 756 | 1.00 | |||||
| Yes | 76 | 2.19 | 1.29, 3.73 | 0.0037 | |||
| Occupation health worker | |||||||
| No | 808 | 1.00 | |||||
| Yes | 24 | 1.03 | 0.45, 2.34 | 0.9487 | |||
| Unemployed | |||||||
| No | 407 | 1.00 | |||||
| Yes | 425 | 0.73 | 0.55, 0.96 | 0.0232 | |||
| HIV status | |||||||
| Negative | 511 | 1.00 | |||||
| Positive | 72 | 1.20 | 0.72, 2.01 | 0.4910 | |||
| Unknown | 249 | 0.60 | 0.44, 0.82 | 0.0012 | |||
| Previous diagnosis of TB | |||||||
| No | 779 | 1.00 | |||||
| Yes | 49 | 1.28 | 0.70, 2.33 | 0.4197 | |||
| Clinical site of disease | |||||||
| Pulmonary | 571 | 1.00 | 0.31, 0.73 | 0.0007 | |||
| Extrapulmonary | 100 | 0.48 | 0.52, 1.06 | 0.0978 | |||
| Both | 161 | 0.74 | |||||
| Chest X-ray lesions | |||||||
| Normal | 70 | 1.00 | |||||
| Cavitary lesions | 163 | 2.34 | 1.32, 4.16 | 0.0037 | |||
| Non-cavitary lesions | 589 | 2.32 | 1.39, 3.85 | 0.0012 | |||
| Treatment outcome | |||||||
| Completed treatment | 668 | 1.00 | |||||
| Died | 69 | 0.97 | 0.59, 1.61 | 0.9154 | |||
| Days of treatment (per 10 days) | 703 | 0.99 | 0.97, 1.00 | 0.0422 | |||
Genotypes were defined by spoligotype and mycobacterial interspersed repetitive units (MIRU) typing.
The univariate logistic regressions included all study cases (N = 832); however, the final multiple regression model included only 825 cases due to cases with missing variable values that had to be excluded.
3.3. Clonal complexes
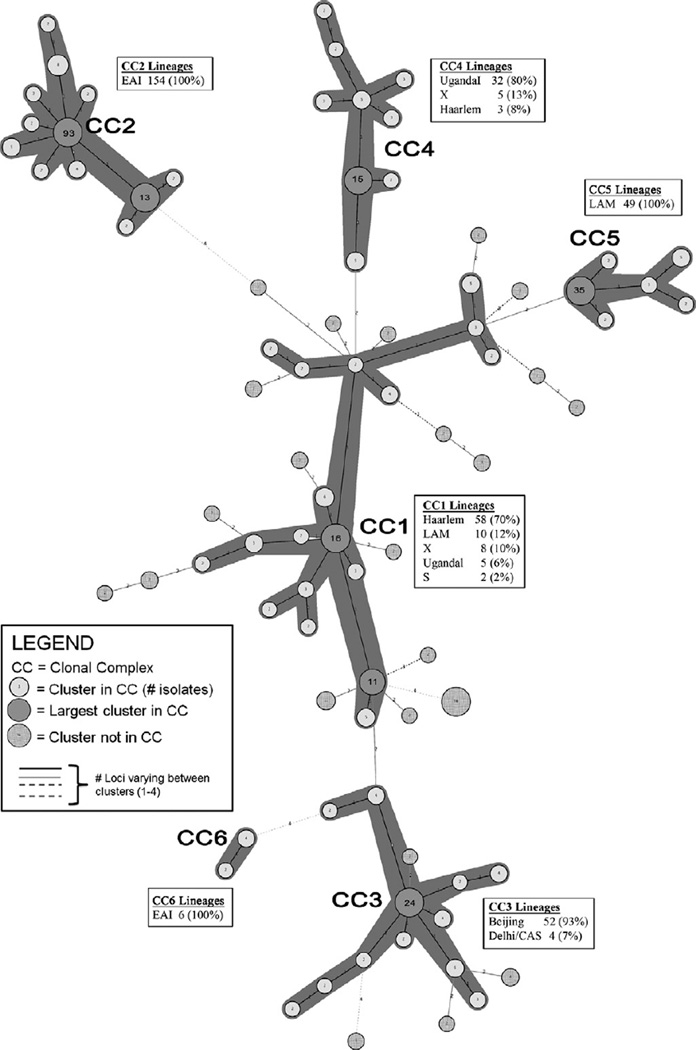
Of the 480 clustered isolates (as determined by exact matching spoligotype and MIRU genotypes – i.e., recently transmitted cases), 388 aggregated into six clonal complexes, with each complex consisting of genetically related strains with no more than one MIRU locus different between them (Fig. 1). Clonal complex group size ranged from 6 isolates (clonal complex 6 [CC6]) to 154 isolates (CC2) (Table 4). There were 92 clustered isolates, representing 25 different MIRU genotypes, that did not sort into a clonal complex because they had more than one MIRU locus different from all other strains analyzed. While CC2 was the largest clonal complex (N = 154 isolates), CC1 was the most diverse, with 21 different genotypes (none differing by more than a single locus).
Fig. 1.
Minimum spanning tree (MST) showing the relationships of MIRU genotypes among 480 clustered Mycobacterium tuberculosis cases in San Diego County (2005–2008). The six clonal complexes (CCs) are demarcated by dark gray shading around circles and represent 388 isolates. The circle nearest to the CC label represents the cluster with the ancestor genotype for that complex. Insert boxes show the number of isolates from different lineages, and proportion of total isolates in each CC.
Table 4.
Characterization of Mycobacterium tuberculosis (Mtb) clonal complexes (CC) found in San Diego County, 2005–2008.
| CC ID | # Mtb strains in CC | # Unique genotypesa in CC | Cluster size (min–max) | Cluster size median |
|---|---|---|---|---|
| CC1 | 83 | 21 | 2–16 | 2 |
| CC2 | 154 | 19 | 2–93 | 2 |
| CC3 | 56 | 12 | 2–24 | 3 |
| CC4 | 40 | 9 | 2–15 | 3 |
| CC5 | 49 | 6 | 2–49 | 3 |
| CC6 | 6 | 2 | 2–4 | 3 |
Genotypes defined by spoligotype and mycobacterial interspersed repetitive units (MIRU) typing.
3.4. Variables associated with clonal complexes
Table 5 shows a frequency analysis of the patient sociodemo-graphic and clinical variables associated with the Mtb strains from CC1 through CC5. CC6 was excluded from the analysis as it had too few (N = 6) isolates. Male patients were more prevalent than females in all clonal complexes, but the proportions differed significantly among the clonal complexes (p = 0.0012). The median age of patients was also significantly different among clonal complexes (p < 0.0001) with patients with CC5 strains having the youngest median age (32.0 years) and patients from CC2 having the oldest median age (54.5 years). While isolates in CC2 came from a few patients of different race/ethnic groups, 85% originated in the Philippines. Furthermore, almost 119/122 (98%) of all the strains that came from TB patients originating in the Philippines were found in CC2. The patients with Mtb strains in CC2 also had low frequencies of non-injection drug use, alcohol abuse, homeless-ness, imprisonments and HIV. CC3 included 16/17 (94%) of the TB patients originating from Vietnam.
Table 5.
Frequency analysisc of socio-demographic and clinic factors associated with clonal complexes identified in San Diego County, 2005–2008 N = 368.
| Variable | Category | Clonal complex (CC) |
X2/Kruskal–Wallis | p | ||||
|---|---|---|---|---|---|---|---|---|
| CC1 (N = 83) | CC2 (N = 154) | CC3 (N = 56) | CC4 (N = 40) | CC5 (N = 49) | ||||
| Socio-demographic risk factors | ||||||||
| Gender | Male | 67 | 81 | 38 | 24 | 70 | 18.14 | 0.0012 |
| Female | 16 | 59 | 18 | 16 | 9 | |||
| Age | Median age in years | 40 | 54.5 | 49.5 | 41 | 32 | 29.14a | <0.0001 |
| Country of birth | Foreign-born | 48 | 130 | 48 | 26 | 31 | 47.83 | <0.0001 |
| US-born | 35 | 10 | 8 | 14 | 18 | |||
| Country of origin | US | 35 | 10 | 8 | 14 | 18 | n/a | |
| Mexico | 39 | 10 | 7 | 26 | 28 | |||
| Philippines | 0 | 119 | 2 | 0 | 1 | |||
| Vietnam | 0 | 1 | 16 | 0 | 0 | |||
| Race/Ethnicity | Asian and native | n/a | ||||||
| Hawaiian | 1 | 121 | 38 | 1 | 1 | |||
| Black | 10 | 1 | 3 | 0 | 2 | |||
| White | 15 | 6 | 5 | 2 | 4 | |||
| Hispanic | 56 | 12 | 10 | 37 | 42 | |||
| Injection drug use | No | 81 | 140 | 56 | 37 | 48 | n/a | |
| Yes | 2 | 0 | 0 | 3 | 1 | |||
| Non-inject drug use | No | 67 | 138 | 52 | 33 | 33 | <0.0001b | |
| Yes | 16 | 2 | 4 | 7 | 16 | |||
| Excessive alcohol use | No | 55 | 132 | 50 | 30 | 39 | 33.13 | <0.0001 |
| Yes | 28 | 8 | 6 | 10 | 10 | |||
| Homeless | No | 66 | 137 | 53 | 35 | 43 | <0.0001b | |
| Yes | 16 | 1 | 3 | 5 | 6 | |||
| Correction facility | No | 71 | 139 | 53 | 33 | 37 | 31.81 | <0.0001 |
| Yes | 12 | 1 | 3 | 7 | 12 | |||
| Unemployed | No | 41 | 74 | 24 | 20 | 23 | 1.75 | 0.7812 |
| Yes | 42 | 66 | 32 | 20 | 26 | |||
| Clinical risk factors | ||||||||
| HIV status | Negative | 53 | 79 | 43 | 24 | 30 | 39.71 | <0.0001 |
| Positive | 13 | 4 | 1 | 9 | 9 | |||
| Unknown | 17 | 57 | 12 | 7 | 10 | |||
| Previous TB disease | No | 78 | 133 | 51 | 35 | 46 | 0.5507b | |
| Yes | 5 | 6 | 5 | 4 | 3 | |||
| Site of disease | Pulmonary | 61 | 102 | 38 | 29 | 39 | 10.20 | 0.2512 |
| Extrapulmonary | 5 | 16 | 10 | 2 | 2 | |||
| Both | 17 | 22 | 8 | 9 | 8 | |||
| X-ray | Cavitary | 19 | 28 | 8 | 6 | 12 | 4.97 | 0.7611 |
| Non-cavitary | 60 | 99 | 42 | 31 | 35 | |||
| Normal | 3 | 11 | 5 | 3 | 2 | |||
| Treatment outcome | Completed Tx | 58 | 111 | 50 | 32 | 29 | n/a | |
| Died | 10 | 17 | 0 | 3 | 2 | |||
| Lost to follow-up | 13 | 10 | 5 | 5 | 8 | |||
| Days of treatment | Median time in days | 195 | 197.5 | 245 | 272.5 | 190 | 12.62 | 0.0133a |
n/a Analysis not appropriate as some cells were zero.
Kruskal–Wallis one-way analysis of variance used.
Fisher exact test used.
Frequencies of categorical variables were compared by X2-test and Fisher’s exact test where noted. Continuous variables were compared by the Kruskal-Wallis one-way analysis of variance where indicated.
All clonal complexes included some strains that came from Hispanic patients, but almost 94% (37/40) of the isolates in CC4 were from Hispanic patients. Both CC4 and CC5 included a high percentage of Hispanics patients from Mexico (93% and 86%, respectively) relative to the other clonal complexes, but CC4 strains came mostly from female patients with a higher median age and CC5 patients had significantly higher proportions of non-injection drug use (33% vs. 17%) and prison admissions compared to CC4 patients (24% vs. 17%).
The proportion of patients with HIV differed significantly among clonal complexes (p < 0.0001). Among the patients tested for HIV, those with Mtb strains from CC4 had the highest prevalence of HIV (27%). The median length of TB treatment of patients in CC3 and CC4 (245 and 272 days, respectively) was one to two months longer than for the other clonal complexes.
4. Discussion
In this study, we examined patient socio-demographic and clinical factors associated with clusters of genotypically related Mtb strains that likely resulted from recent person-to-person transmission events. We examined factors related to clusters of identical genotypes as well as factors related to clonal clusters of closely related strains.
Of the 832 San Diego TB cases evaluated from 2005 through 2008, the majority of patients (58%) had Mtb isolates that had identical spoligotype and MIRU-12 genotypes to at least one other isolate. These results indicate that almost 50% of TB cases in this region likely resulted from recent transmission while the other half were probably the result of reactivation events, suggesting that enhanced local and international TB control will be needed to manage TB in binational regions such as San Diego. It is important to note, however, that we used a combination of MIRU-12 genotyping plus spoligotyping to determine if two TB isolates had identical genotypes. While this was the genotyping standard in the US until recently, it has now been demonstrated that genotyping by MIRU-12 plus spoligotyping could overestimate the cluster proportion in a cosmopolitan set of isolates by between 20% and 30% (Maes et al., 2008; Supply et al., 2006). Considering this likely over-estimation of clustering based on MIRU-12 genotyping, we estimate that the true cluster proportion in this region is probably closer to 40–46%. Either way, the cluster proportion (40–58%) found in San Diego, is well within the range of cluster proportions described in a recent systematic review of global TB genotyping studies (global cluster proportion range: 7–72.3%). However, even a conservative estimate of the San Diego cluster proportion is above the global mean (38.7% clustering), suggesting a higher than average recent transmission of TB in this region. The San Diego case cluster proportion is also higher than most of the other studies of TB case clustering in the US that used the same definition of identical genotypes as a standard (Bishai et al., 1998; Burman et al., 1997; de Bruyn et al., 2001; Ellis et al., 2002; Geng et al., 2002; Kempf et al., 2005; Rhee et al., 2000). It is notable that the only US study in the above mentioned systematic review that demonstrated a higher cluster proportion than San Diego, was conducted in Houston, Texas (60.5%) (de Bruyn et al., 2001), another region with high immigration. Improved strategies for screening non-immigrant entrants such as students and temporary workers, and assuring resources remain in place for helping high incidence countries perform adequate contact screening and treatment, will be critical to long-term prevention of TB in high immigration regions of the US.
In this study, we observed that patients whose Mtb isolates clustered by genotype were significantly more likely to be young, male and homeless than those with singleton genotypes, which supports the hypothesis that these infections were recent. Home-lessness is well known to be associated with TB case clustering in regions of low TB incidence globally, but not in high incidence countries (Fok et al., 2008). While substantial effort already goes into preventing TB in homeless shelters in San Diego, these data suggest that more aggressive prevention efforts and more active case finding focused on this cohort might help reduce the incidence of recent transmission TB in San Diego. Our data also indicates that patients born in the Philippines were 1.86 times as likely to have clustered Mtb isolates compared to those born in the US, which suggests that recent transmission may play a larger role in this patient population than others in San Diego.
While almost half of the cases analyzed likely originated from recent transmission events, the Mtb strains in San Diego represent many diverse and globally prevalent lineages (Brudey et al., 2006). Further study would be needed to determine whether these strains came into the region via immigration and then were propagated locally, or whether they merely reflect the prevalence of these strains in the countries from which the patients immigrated. It is notable that the Beijing and S lineages were more than twice as prevalent in the clustered cases compared to the singleton cases, and X lineage strains were more common in singleton cases, suggesting possible associations between Beijing and S lineages and recent transmission. Recent studies have suggested that these strains are more virulent, and associated with worse clinical disease (Feng et al., 2009), high treatment failure rates (Parwati et al., 2010) and drug resistance (Drobniewski et al., 2005), which might explain the prevalence of this lineage in recent transmission cases. However, it is also important to note that Beijing strains can be poorly differentiated by MIRU-15 genotyping (Luo et al., 2012), which is more discriminatory than the MIRU-12 typing we used. Clustering of strains in this lineage in our study may therefore have been overestimated and should be interpreted with caution.
Clonal complexes in this analysis were conservatively defined as groups of clustered Mtb strains with up to one MIRU loci difference between them, thereby presumably representing a group of genetically related strains (Allix-Beguec et al., 2008; Weniger et al., 2010). While the TB patients with strains in the same clonal complex had some overlapping sociodemographic and clinical features, they tended to have unique racial/ethnic characteristics. Specifically, CC1 was made up of strains from mostly Hispanic and white patients born in the US, while CC2 included almost all the Philippines patients and CC3 was made up mostly of patients originating in Vietnam. CC4 and CC5 were made up mostly of Hispanics born in Mexico.
There is overlap, however. CC2 for example, while mostly made up of strains derived from Asian patients from the Philippines, also includes Mtb strains collected from Hispanic, White and Black patients from the US and Mexico. The most parsimonious explanation for this socio-demographic distribution of isolates would be that the strains from Filipino patients for example, (which make up 85% of CC2) were originally brought into San Diego by immigration and then transmitted to White, Black and Hispanic sub-populations in San Diego. While the reverse could also be true, it is less likely as all isolates in this CC are of the EAI lineage, which is found predominantly in patients from East Asia. What this demonstrates is that among the most recently transmitted strains, clonal complexes of related Mtb strains are staying largely confined within the sub-populations of patients in which the strains likely originated. One possible explanation for this is that local transmission of TB in San Diego is occurring largely within social networks defined by ethnicity. However, a detailed temporal and epidemiological study of the isolates in the clonal complexes, which is beyond the scope of this study, is needed to examine this question in more detail.
There were significantly different proportions of HIV positive patients in the different clonal complexes, with patients harboring CC4 and CC5 strains having the highest prevalence of HIV (27% and 23% of those tested). A recent multivariate analysis of HIV-TB cases in San Diego, which found HIV-TB significantly associated with Hispanic ethnicity (Rodwell et al., 2010a), suggests the observed association between CC4 and CC5, and HIV prevalence, is likely confounded by the high proportion of Hispanics in CC4 and CC5 (93% and 86% respectively), rather than anything to do with the Mtb strains in CC4. Other significant socio-demographic factors associated with clonal complexes, for example, homelessness and non-injection drug use are likely similarly confounded by the high proportions of specific ethnicities in each clonal complex.
At a population level, our findings suggest that categorizing Mtb genotypes into clonal complexes could have public health implications that go beyond the evaluation of cluster proportions and person-to-person contact tracing. Given the dominance of specific races and ethnicities of patients within related Mtb clonal complexes, it would seem that race/ethnicity appropriate and targeted prevention strategies – prioritized by dominance of the clonal complex in the region – could be utilized to attempt eradication of TB in the region, one clonal complex at a time.
5. Limitations
Only MIRU-12 typing data was available for this study, and it is known to be less discriminatory than MIRU-24 typing. It is therefore possible that we overestimated clustering proportions in this study by as much as 20–30% as explained in the Discussion section. It is possible for epidemiologically unrelated Mtb strains to evolve the same genotype independently, a phenomenon known as homoplasy. If homoplasy were occurring in the Mtb strains in San Diego, then we could have misclassified some strains as related when they were in fact not. If this occurred, it would have most likely decreased the strength of our reported associations. In clonal organisms such as Mtb, however, homoplasy is extremely rare. As most Mtb genotyping methods are based on observing only a small fraction of the total genome, it is also possible that two unrelated strains could appear related due to homology in the section of the genome observed (many genes are conserved in Mtb). The likelihood of this is limited by characterizing multiple regions of the pathogen genome, which we did by using both spoligotyping and MIRU data to define the genotype of each isolate. As most of the Mtb strains examined in this study were isolated from patients born outside the US, we compared all strains to the globally representative SpolDB4 database to determine lineage and “orphan” status of the strains. It is possible that strains designated as orphans in this study may have matched to other existing lineages if they were compared to additional existing strain databases from the US and elsewhere.
6. Conclusion
Patients with clustered Mtb isolates that were stratified into clonal complexes of related strains had distinctive and largely unique socio-demographic and clinical characteristics. These data suggest TB in this region is being transmitted in relatively insular community networks defined by race/ethnicity and country of origin. The addition of clonal complex analysis to simple cluster analysis provides important public health insights into the transmission of TB in multinational regions with diverse Mtb genotypes.
Supplementary Material
Acknowledgements
T.C. Rodwell was supported by career development Grant K01AI083784 from the National Institute of Allergy and Infectious Disease.
We wish to acknowledge Dr. Marisa Moore for her help with data collection and extraction and Dr. Ed Desmond and Grace Lin for all their hard work genotyping the San Diego TB strains.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.meegid.2012.08.022.
Contributor Information
Timothy C. Rodwell, Email: trodwell@ucsd.edu.
Anokhi J. Kapasi, Email: anokhik@gmail.com.
Richard F.W. Barnes, Email: rgarfein@ucsd.edu, r1barnes@ucsd.edu.
Kathleen S. Moser, Email: Kathleen.Moser@sdcounty.ca.gov.
References
- Allix-Beguec C, Harmsen D, Weniger T, Supply P, Niemann S. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J. Clin Microbiol. 2008;46:2692–2699. doi: 10.1128/JCM.00540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr MA, Mostowy S. Molecular tools for typing and branding the tubercle bacillus. Curr. Mol. Med. 2007;7:309–317. doi: 10.2174/156652407780598593. [DOI] [PubMed] [Google Scholar]
- Bezanahary H, Baclet MC, Sola C, Gazaille V, Turlure P, Weinbreck P, Denis F, Martin C. Molecular strain typing contribution to epidemiology of tuberculosis in Limousin (1998 to 2006) Med. Mal. Infect. 2008;38:309–317. doi: 10.1016/j.medmal.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Bishai WR, Graham NMH, Harrington S, Pope DS, Hooper N, Astemborski J, Sheely L, Vlahov D, Glass GE, Chaisson RE. Molecular and geographic patterns of tuberculosis transmission after 15 years of directly observed therapy. JAMA. 1998;280:1679–1684. doi: 10.1001/jama.280.19.1679. [DOI] [PubMed] [Google Scholar]
- Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, Allix C, Aristimuno L, Arora J, Baumanis V, Binder L, Cafrune P, Cataldi A, Cheong S, Diel R, Ellermeier C, Evans JT, Fauville-Dufaux M, Ferdinand S, Garcia de Viedma D, Garzelli C, Gazzola L, Gomes HM, Guttierez MC, Hawkey PM, van Helden PD, Kadival GV, Kreiswirth BN, Kremer K, Kubin M, Kulkarni SP, Liens B, Lillebaek T, Ly HM, Martin C, Martin C, Mokrousov I, Narvskaia O, Ngeow YF, Naumann L, Niemann S, Parwati I, Rahim Z, Rasolofo-Razanamparany V, Rasolonavalona T, Rossetti ML, Ruesch-Gerdes S, Sajduda A, Samper S, Shemyakin IG, Singh UB, Somoskovi A, Skuce RA, van Soolingen D, Streicher EM, Suffys PN, Tortoli E, Tracevska T, Vincent V, Victor TC, Warren RM, Yap SF, Zaman K, Portaels F, Rastogi N, Sola C. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpoIDB4) for classification, population genetics and epidemiology. BMC. Microbiol. 2006;6:23. doi: 10.1186/1471-2180-6-23. http://dx.doi.org/10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman WJ, Reves RR, Hawkes AP, Rietmeijer CA, Yang ZH, ElHajj H, Bates JH, Cave MD. DNA fingerprinting with two probes decreases clustering of Mycobacterium tuberculosis. Am. J. Respir. Crit. Care Med. 1997;155:1140–1146. doi: 10.1164/ajrccm.155.3.9117000. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Case definitions for infectious conditions under public health surveillance. MMWR Recomm. Rep. 1997;46:1–55. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) [accessed 20.11.2010];Guide to the Application of Genotyping to Tuberculosis Preventiona and Control. 2004 Available at: < http://www.cdc.gov/tb/programs/genotyping/manual.htm>.
- Centers for Disease Control and Prevention (CDC) Trends in tuberculosis – United States, 2010. MMWR Morb. Mortal. Wkly. Rep. 2011;60:333–337. [PubMed] [Google Scholar]
- Cowan LS, Mosher L, Diem L, Massey JP, Crawford JT. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J. Clin. Microbiol. 2002;40:1592–1602. doi: 10.1128/JCM.40.5.1592-1602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan LS, Diem L, Brake MC, Crawford JT. Transfer of a Mycobacterium tuberculosis genotyping method, spoligotyping, from a reverse line-blot hybridization, membrane-based assay to the Luminex multianalyte profiling system. J. Clin. Microbiol. 2004;42:474–477. doi: 10.1128/JCM.42.1.474-477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale JW, Brittain D, Cataldi AA, Cousins D, Crawford JT, Driscoll J, Heersma H, Lillebaek T, Quitugua T, Rastogi N, Skuce RA, Sola C, Van Soolingen D, Vincent V. Spacer oligonucleotide typing of bacteria of the Mycobacterium tuberculosis complex: recommendations for standardised nomenclature. Int. J. Tuberc. Lung Dis. 2001;5:216–219. [PubMed] [Google Scholar]
- David S. Strategical use of genotyping of Mycobacterium tuberculosis in tuberculosis control. Rev. Port. Pneumol. 2008;14:509–516. doi: 10.1016/s0873-2159(15)30255-5. [DOI] [PubMed] [Google Scholar]
- de Bruyn G, Adams GJ, Teeter LD, Soini H, Musser JM, Graviss EA. The contribution of ethnicity to Mycobacterium tuberculosis strain clustering. Int. J. Tuberc. Lung Dis. 2001;5:633–641. [PubMed] [Google Scholar]
- Drobniewski F, Balabanova Y, Nikolayevsky V, Ruddy M, Kuznetzov S, Zakharova S, Melemyev A, Fedorin I. Drug-resistant, tuberculosis, clinical virulence, and the dominance of the Beijing strain family in Russia. JAMA. 2005;293:2726–2731. doi: 10.1001/jama.293.22.2726. [DOI] [PubMed] [Google Scholar]
- Ellis BA, Crawford JT, Braden CR, McNabb SJN, Moore M, Kammerer S. Natl Tuberculosis Genotyping Molecular epidemiology of tuberculosis in a sentinel surveillance population. Emerg. Infect. Dis. 2002;8:1197–1209. doi: 10.3201/eid0811.020403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng JY, Su WJ, Liu LY, Tsai CC, Chang SC. Radiological presentation of pulmonary tuberculosis infected by the W-Beijing Mycobacterium tuberculosis strain. Int. J. Tuberc. Lung Dis. 2009;13:1387–1392. [PubMed] [Google Scholar]
- Fica A, Cifuentes M, Ajenjo MC, Jemenao MI, Zambrano A, Febre N, Delpiano L, Diomedi A, Ramonda P, Com Consultivo I. Tuberculosis in healthcare workers. Rev. Chilena Infectol. 2008;25:243–255. [PubMed] [Google Scholar]
- Fok A, Numata Y, Schulzer M, FitzGerald MJ. Risk factors for clustering of tuberculosis cases: a systematic review of population-based molecular epidemiology studies. Int. J. Tuberc. Lung Dis. 2008;12:480–492. [PubMed] [Google Scholar]
- Garzelli C, Rindi L. Molecular epidemiological approaches to study the epidemiology of tuberculosis in low-incidence settings receiving immigrants. Infect. Genet. Evol. 2012;12:610–618. doi: 10.1016/j.meegid.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Geng E, Kreiswirth B, Driver C, Li JH, Burzynski J, DellaLatta P, LaPaz A, Schluger NW. Changes in the transmission of tuberculosis in New York City from 1990 to 1999. N. Engl. J. Med. 2002;346:1453–1458. doi: 10.1056/NEJMoa012972. [DOI] [PubMed] [Google Scholar]
- Gibson AL, Huard RC, van Pittius NCG, Lazzarini LCO, Driscoll J, Kurepina N, Zozio T, Sola C, Spindola SM, Kritski AL, Fitzgerald D, Kremer K, Mardassi H, Chitale P, Brinkworth J, de Viedma DG, Gicquel B, Pape JW, van Soolingem D, Kreiswirth BN, Warren RM, van Helden PD, Rastogi N, Suffys PN, Silva JLE, Ho JL. Application of sensitive and specific molecular methods to uncover global dissemination of the major RDRio sublineage of the Latin american-mediterranean Mycobacterium tuberculosis spoligotype family. J. Clin. Microbiol. 2008;46:1259–1267. doi: 10.1128/JCM.02231-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange JM, Yates MD, de Kantor IN. Guidelines for Speciation within the Mycobacterium tuberculosis Complex. 1996 Available at: < http://www.whqlibdoc.who.int/hq/1996/WHO_EMC_ZOO_96.4.pdf> (WHO/EMC/ZOO/96.4)
- International Community Foundation (ICF) [accessed 25.01.2011];Tuberculosis in the San Diego– Tijuana Border Region: Inspiring Philanthropy Beyond Borders. 2010 Available at: < http://www.icfdn.org/reports/tbstudy/>.
- Kamerbeek J, Schouls L, Kolk A, vanAgterveld M, vanSoolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, vanEmbden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf MC, Dunlap NE, Lok KH, Benjamin WH, Keenan NB, Kimerling ME. Long-term molecular analysis of tuberculosis strains in Alabama, a state characterized by a largely indigenous, low-risk population. J. Clin. Microbiol. 2005;43:870–878. doi: 10.1128/JCM.43.2.870-878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange JE, Lauer EM, Voas RB. A survey of the San Diego-Tijuana cross-border binging: methods and analysis. Eval. Rev. 1999;23:378–398. doi: 10.1177/0193841X9902300402. [DOI] [PubMed] [Google Scholar]
- Luo T, Yang C, Gagneux S, Gicquel B, Mei J, Gao Q. Combination of single nucleotide polymorphism and variable-number tandem repeats for genotyping a homogenous population of Mycobacterium tuberculosis Beijing strains in China. J. Clin. Microbiol. 2012;50:633–639. doi: 10.1128/JCM.05539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes M, Kremer K, van Soolingen D, Takiff H, de Waard JH. 24-locus MIRU-VNTR genotyping is a useful tool to study the molecular epidemiology of tuberculosis among Warao Amerindians in Venezuela. Tuberculosis. 2008;88:490–494. doi: 10.1016/j.tube.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Mathema B, Kurepina N, Fallows D, Kreiswirth BN. Lessons from molecular epidemiology and comparative genomics. Semin. Respir. Crit. Care Med. 2008;29:467–480. doi: 10.1055/s-0028-1085699. [DOI] [PubMed] [Google Scholar]
- Mazars E, Lesjean S, Banuls AL, Gilbert M, Vincent V, Gicquel B, Tibayrenc M, Locht C, Supply P. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis . Proc. Natl. Acad. Sci. USA. 2001;98:1901–1906. doi: 10.1073/pnas.98.4.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe JZ, Kim EY, Lin SY, Cattamanchi A, Oh P, Flood J, Hopewell PC, Kato-Maeda M. Determinants of multidrug-resistant tuberculosis clusters, California, USA, 2004–2007. Emerg. Infect. Dis. 2010;16:1403–1409. doi: 10.3201/eid1609.100253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parwati I, Alisjahbana B, Apriani L, Soetikno RD, Ottenhoff TH, van der Zanden AGM, van der Meer J, van Soolingen D, van Crevel R. Mycobacterium tuberculosis Beijing genotype is an independent risk factor for tuberculosis treatment failure in Indonesia. J. Infect. Dis. 2010;201:553–557. doi: 10.1086/650311. [DOI] [PubMed] [Google Scholar]
- Rhee JT, Tanaka MM, Behr MA, Agasino CB, Paz EA, Hopewell PC, Small PM. Use of multiple markers in population-based molecular epidemiologic studies of tuberculosis. Int. J. Tuberc. Lung Dis. 2000;4:1111–1119. [PubMed] [Google Scholar]
- Rodwell TC, Moore M, Moser KS, Brodine SK, Strathdee SA. Tuberculosis from Mycobacterium bovis in binational communities, United States. Emerg. Infect. Dis. 2008;14:909–916. doi: 10.3201/eid1406.071485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodwell TC, Barnes RFW, Moore M, Strathdee SA, Raich A, Moser KS, Garfein RS. HIV–Tuberculosis coinfection in southern California: evaluating disparities in disease burden. Am. J. Public Health. 2010a;100:S178–S185. doi: 10.2105/AJPH.2009.170142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodwell TC, Kapasi AJ, Moore M, Milian-Suazo F, Harrise B, Guerrerod LP, Moser KS, Strathdee SA, Garfein RS. Tracing the origins of Mycobacterium bovis tuberculosis in humans in the United States to cattle in Mexico using spoligotyping. Int. J. Infect. Dis. 2010b;14S:e129–e135. doi: 10.1016/j.ijid.2009.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- San Diego Health Human Service Agency (HHSA) Comparison of National, State and County Data for Tuberculosis, 2003–2010. 2011 Available at: http://www.sdcounty.ca.gov/hhsa/programs/phs/documents/fctshttables2010V1.pdf.
- Small PM, Hopewell PC, Singh SP, Paz A, Parsonnet J, Ruston DC, Schecter GF, Daley CL, Schoolnik GK. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- Stokes ME, Davis CS, Koch GG. Categorical Data Analysis Using the Sas System. second ed. Cary, North Carolina: SAS Institute Inc.; 2000. [Google Scholar]
- Streicher EM, Victor TC, van der Spuy G, Sola C, Rastogi N, van Helden PD, Warren RM. Spoligotype signatures in the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 2007;45:237–240. doi: 10.1128/JCM.01429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis . J. Clin. Microbiol. 2006;44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Census Bureau. US Census Bureau: State and county quickfacts. 2011 Available at: < http://www.quickfacts.census.gov/qfd/states/06/06073.html>.
- Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 2010;38:W326–W331. doi: 10.1093/nar/gkq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar JH. Biostatistical Analysis. fourth ed. Upper Saddle River: Prentice Hall Inc.; 1999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.