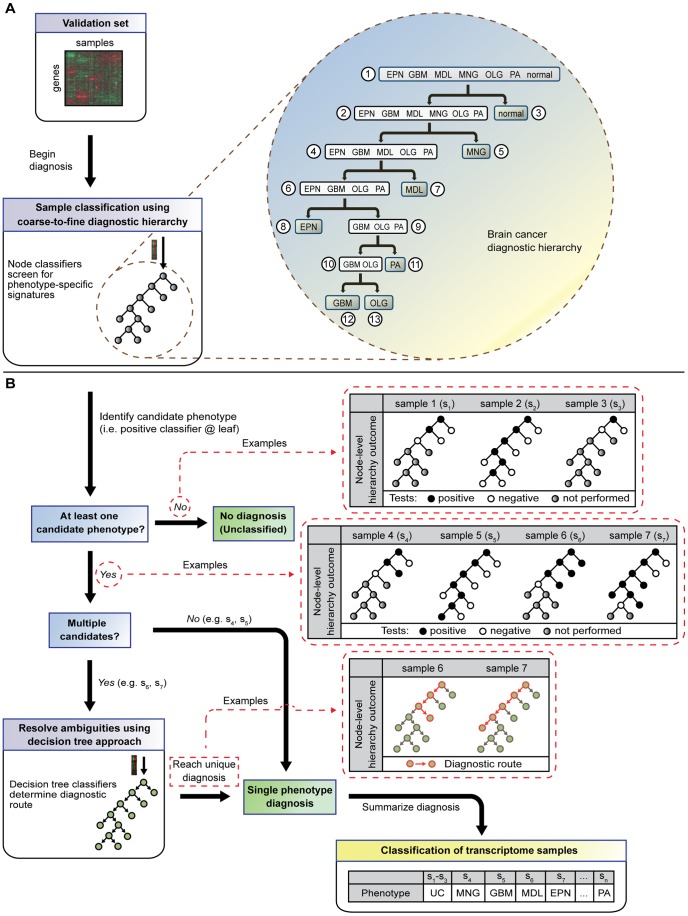
Figure 3. Comprehensive classification of human brain cancer and normal brain transcriptomes using molecular signatures from ISSAC.
A The coarse-to-fine classification process is represented by a hierarchically structured groupings of phenotypes. There is a node classifier for each set of phenotypes in the hierarchy, which is designed to respond positively if the sample belongs to this set of diseases and negatively otherwise. Our diagnostic hierarchy has thirteen nodes in total, and seven terminal nodes (i.e., leaves). The node classifiers are executed sequentially and adaptively on a given expression profile; a classifier test for a particular node is performed if and only if all of its ancestor tests were performed and deemed positive. The node classifiers are used to screen for phenotype-specific signatures. B Leaves that have positive classifier outcomes correspond to the candidate phenotypes of a given expression profile. If there is no candidate phenotype, the expression profile is labeled as ‘Unclassified’. If only one candidate phenotype is identified, the profile is labeled as that phenotype of the respective leaf. If the profile is considered to consist of multiple phenotype signatures, the ambiguity is resolved using the decision-tree classifiers based on the same diagnostic hierarchy. Here, the decision-tree classifiers are executed starting from the root of the tree, directing the profile to one of the two child nodes sequentially until it completes a full path towards a leaf. The phenotype label of the final destination corresponds to the unique diagnosis.