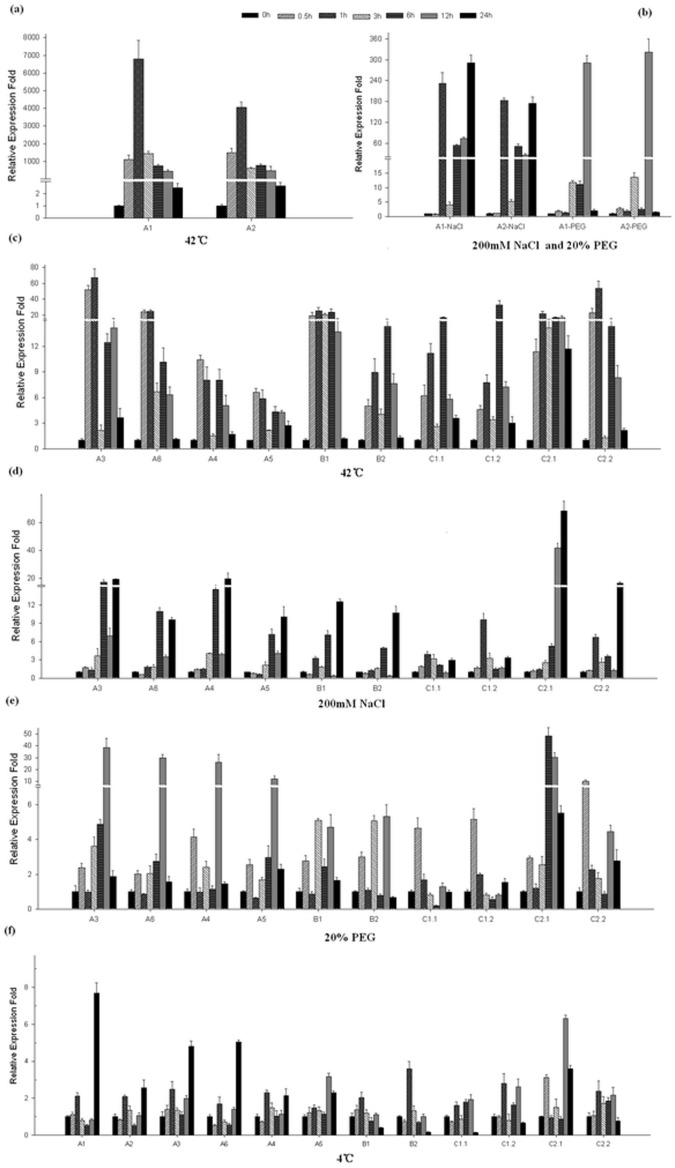
Figure 3. Quantitative real-time PCR analyses of the relative expression fold of 12 GmHsp90 genes under different stresses in soybean.
Leaves of soybean from all the treatments harvested in 0, 0.5, 1, 3, 6, 12 and 24 h were used for the analysis. A soybean beta tubulin gene was used as internal control for normalization and the relative mRNA level for each gene was calculated as ΔΔCT values. The result of expression was processed as relative expression fold and was generated by SigmaPlot 9.0. The transcript levels (means ± SD) displayed were each calculated using the qRT- PCR results of three technical repeats. Leaves of each repeat were prepared from at least five independent plants. (a) Relative expression folds of GmHsp90A1 and GmHsp90A2 under heat shock. (b) Relative expression folds of GmHsp90A1 and GmHsp90A2 under salt and osmotic stress. (c) Relative expression folds of the remainder of GmHsp90 genes under heat stress. (d) Relative expression folds of the remainder of GmHsp90 genes under salt stress. (e) Relative expression folds of the remainder of GmHsp90 genes under osmotic tress. (f) Relative expression folds of GmHsp90 genes under cold stress. A1, GmHSP90A1; A2, GmHSP90A2; A3, GmHSP90A3; A4, GmHSP90A4; A5, GmHSP90A5; A6, GmHSP90A6; B1, GmHSP90B1; B2, GmHSP90B2; C1.1, GmHSP90C1.1; C1.2, GmHSP90C1.2; C2.1, GmHSP90C2.1; C2.2, GmHSP90C2.2.