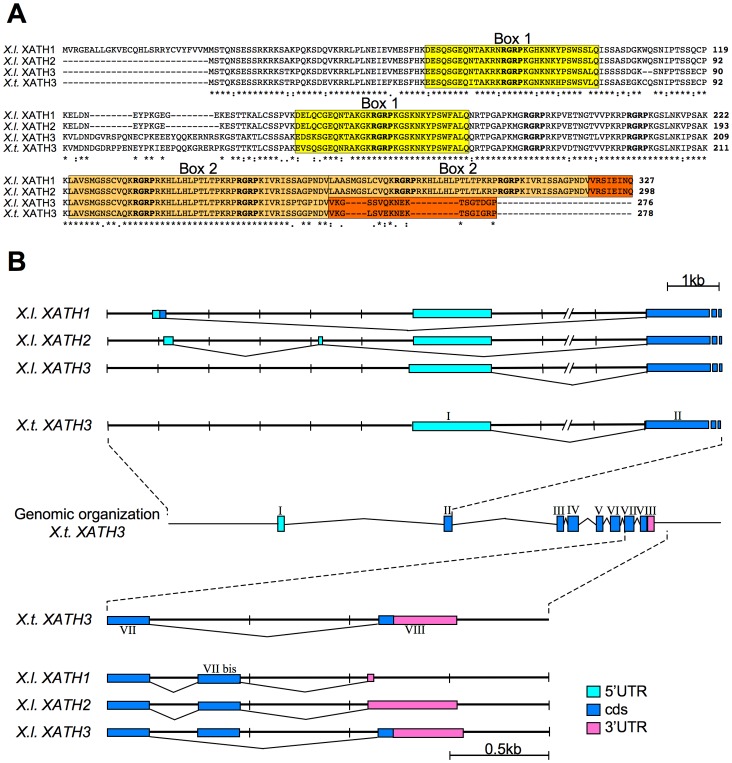
Figure 1. XHMG-AT-hook proteins and organization of their transcripts and loci.
(A) ClustalW alignment of XHMG-AT-hook protein isoforms. The amino acid sequences of the three different XHMG-AT-hook1-3 protein sequences (XATH1–3) found in X. laevis and of the one (XATH3) found in X. tropicalis are shown. The conserved AT-hooks are shown in bold; internal repeats are boxed in different shades of yellow or brown respectively. The C-terminal region is boxed in orange. (B) Genomic organization of the Xhmg-at-hook locus in Xenopus tropicalis. The exon/intron organization is indicated together with the proposed mechanisms of generation of the different Xhmg-at-hook1-3 (XATH1-3) transcripts in Xenopus laevis, based on homology with the genomic sequences of Xenopus tropicalis (see also description in the text).