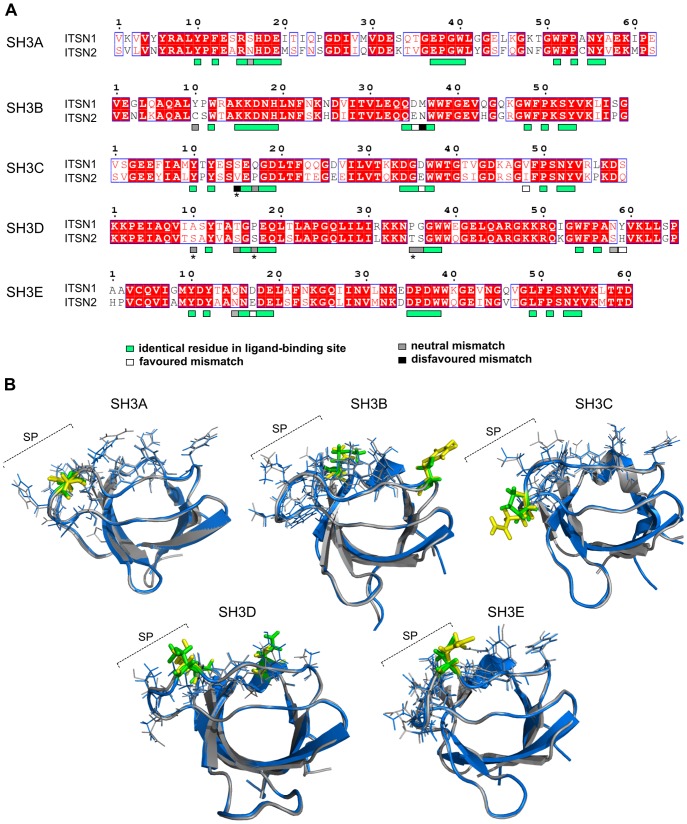
Figure 2. Comparison of ligand-binding sites of the SH3 domains of ITSN1 and ITSN2.
(A) Alignment of protein sequences of the human ITSNs SH3 domains. The alignment was generated using the ClustalW algorithm. Identical residues are highlighted in red, and homologous amino acids are shown in red letters. Amino acid residues that form ligand-binding sites of the SH3 domains are indicated by boxes, mismatches within these regions are shown by boxes of different colours. Amino acid residues marked with an asterisk are not conserved in ITSN1 and ITSN2 orthologues. (B) Merged structures are shown for each SH3 domain pair in blue and grey for ITSN1 and ITSN2, respectively. Amino acid residues that form ligand-binding sites of the SH3 domains and are identical in ITSNs or not conserved in orthologues are depicted as lines. Divergent residues involved in ligand binding of the ITSN1 and ITSN2 SH3 domains are depicted in yellow and green, respectively. Amino acid mismatches regarded as favoured are not shown in colour. SP, specificity pocket of the SH3 domain ligand-binding site.