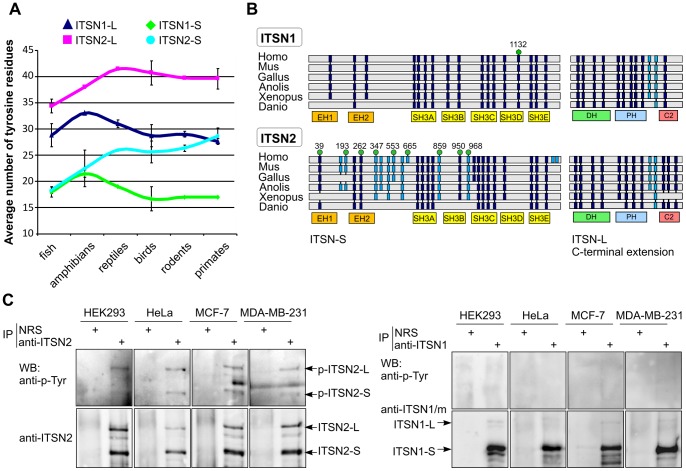
Figure 4. ITSN2 is phosphorylated on tyrosine residues.
(A) Line graphs show the total number of tyrosine residues in ITSN1 and ITSN2 in a range of vertebrate taxonomic groups. Amino acid sequences of ITSN1 and ITSN2 orthologues in fish (Danio rerio, Oreochromis niloticus and Tetraodon nigroviridis), amphibians (Xenopus tropicalis and Xenopus laevis), reptiles (Anolis carolinensis), birds (Gallus gallus, Meleagris gallopavo and Taeniopygia guttata), rodents (Mus musculus, Rattus norvegicus and Cricetulus griseus) and primates (Homo sapiens, Pan troglodita and Pongo abelii) were obtained from the NCBI protein database (http: //ncbi.nlm.nih.gov). The average number of tyrosine residues per taxon is plotted, and error bars represent standard deviations. (B) Schematic representation of ITSNs domain organization and distribution of conserved tyrosine residues in ITSN1 and ITSN2 of various vertebrates. Tyrosine residues located within domain and interdomain regions are shown as dark blue and light blue boxes, respectively. Tyrosine residues that could be phosphorylated according to phosphoproteomic data (www.phosphosite.org) are indicated by green circles. The number above each circle indicates the position of the residue in the amino acid sequence of human ITSN. Abbreviations are defined as follows: Homo, Homo sapiens; Mus, Mus musculus; Gallus, Gallus gallus; Anolis, Anolis carolinensis; Xenopus, Xenopus laevis. (C) Lysates of growing HEK293, HeLa, MCF-7 and MBA-MB-231 cells were subjected to immunoprecipitation with rabbit polyclonal anti-ITSN2 (left panel) or anti-ITSN1 (right panel) antibodies. Normal rabbit serum (NRS) was used as control. Precipitated proteins were analyzed by Western blotting using anti-phosphotyrosine antibodies and antibodies against ITSN1 or ITSN2. The experiments were repeated at least twice with reproducible results. IP, immunoprecipitation; WB, Western blotting.