Figure 5.
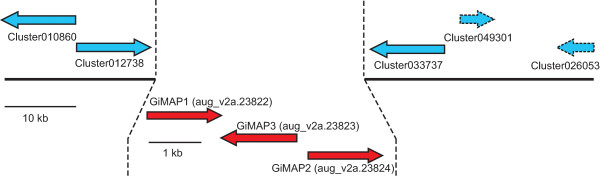
Organisation of the coral GiMAP loci. As shown in the upper part of the figure (in blue) the genomic context of the GiMAP loci is the same in both A. millepora and A. digitifera. Numbers on the arrows refer to the A. millepora transcriptome assembly [20]. Cluster010860 encodes a protein phosphatase regulatory subunit, and corresponds to the A. digitifera predicted protein aug_v2a.23820.t1. Cluster012738 encodes a lysosomal acid lipase, and corresponds to A. digitifera aug_v2a.23821.t1. Cluster033737 corresponds to aug_v2a.23825.t1, Cluster049301 corresponds to the 3′ end of aug_v2a.23826.t1 and Cluster026053 corresponds to aug_v2a.23827.t1; all lack clear orthologs in the database. As EST support is lacking, the orientation of clusters 049301 and 026053 in A. millepora could not be confirmed; the orientation is assumed to be as in A. digitifera, but is shown in broken outline to reflect the uncertainty. As shown in the lower part of the figure (in red) the A. digitifera orthologs of the three A. millepora GiMAP genes are organised in a tight cluster. In the case of A. millepora, the corresponding transcriptome clusters map to a region of similar size, but the organisation of the loci could not be unequivocally established. For A. millepora, Cluster012256 = GiMAP1, Cluster 024980 = GiMAP2 and Cluster015015 = GiMAP3) Note that the GiMAP loci are also tightly linked in mammals and plants, but are the products of independent lineage-specific expansions.
