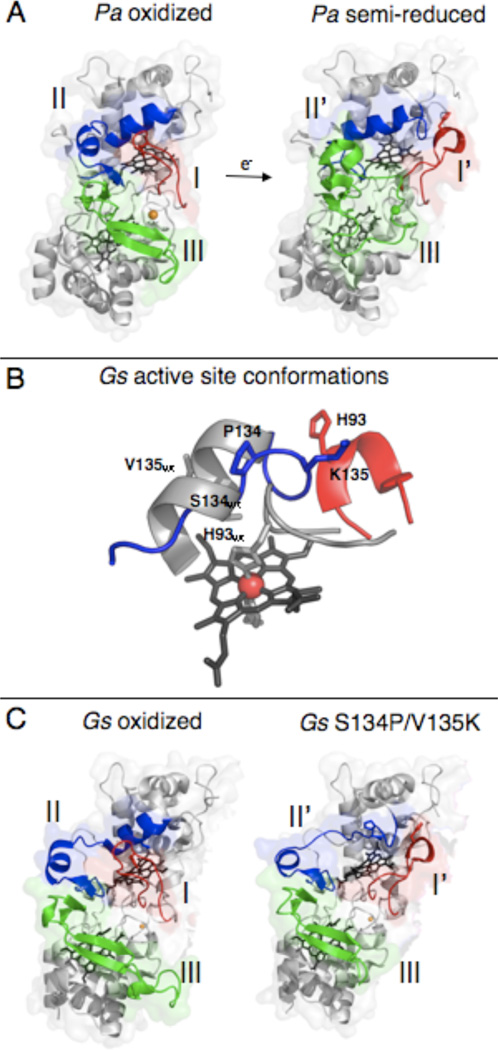
Figure 1.
Crystal structures of members of the canonical class of bacterial cytochrome c peroxidases. (A) Structures of the oxidized P. aeruginosa CcP (top, PDB 1EBY) with loops I (red), II (blue), and III (green) highlighted. The addition of one electron to reduce the high potential heme causes the rearrangement of loops I, II, and III in the semi-reduced protein (bottom, PDB 2VHD). (B) G. sulfurreducens in both the wild-type oxidized (grey) and S134P/V135K mutant (red/blue) enzyme. The mutations to residues S134 and V135 in the wild-type protein has significant effects on the structure surrounding the distal face of the L-heme. This changes the activity requirements of the enzyme seen in solution and PFV analyses. (Figures of the active site of wild-type and S134P/V135K were prepared using Protein Data Bank files 3HQ6 and 3HQ8 respectively (15).) (C) Structures of the wild-type and double mutant S134P/V135K enzymes from G. sulfurreducens. The wild-type oxidized protein (top, PDB 3HQ6) has loop I (red) in the closed position. The S134P and V135K mutations (bottom, PDB 3HQ8) cause only loops I (red) and II (blue) to rearrange. Loop III (green) retains the wild-type conformation and does not resemble the semi-reduced form of the P. aeruginosa enzyme.