Figure 2.
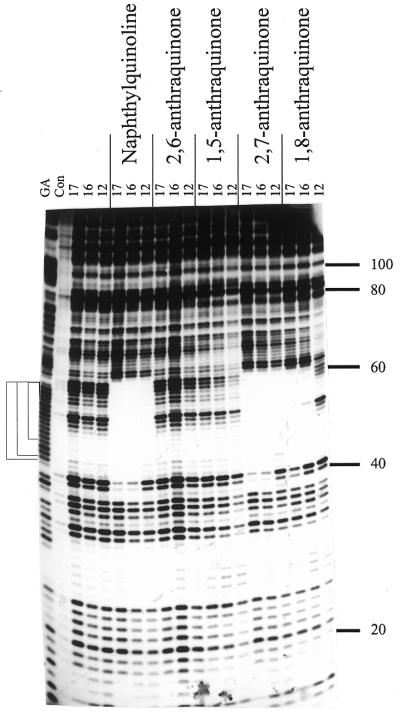
DNase I footprints for the interaction of the various TG-oligonucleotides (10 µM) in the presence of 10 µM of the triplex-binding ligands. The length of the oligonucleotide is shown at the top of each gel lane. The track labelled ‘Con’ shows DNase I digestion of the duplex DNA in the absence of oligonucleotide or ligand. Note that this lane is underdigested in this figure, but the cleavage pattern is identical to the adjacent lanes containing oligonucleotides with no added ligand. ‘GA’ represents a Maxam–Gilbert marker specific for purines. The experiments were performed in 10 mM Tris–HCl pH 7.0 containing 50 mM NaCl and 10 mM MgCl2. The numbers on the right-hand side indicate the sequence position and are numbered as in Brown et al. (25). The square brackets show the position of the expected target sites.
