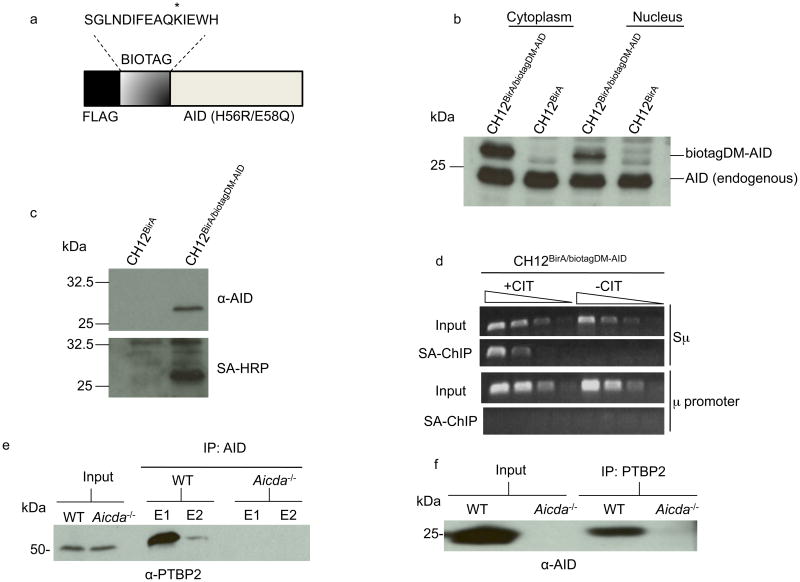
Figure 1.
AID interacts with PTBP2. (a) Schematic representation of the AID expression construct (biotagDM-AID). The lysine that is biotinylated by BirA is indicated with an asterisk. The H56R,E58Q mutation inactivates the DNA deaminase activity of AID. (b) Protein extracts derived from stimulated CH12BirA or CH12BirA/biotagDM-AID cells were analyzed on immunoblots with AID antibodies. (c) Cell extracts from CH12BirA or CH12BirA/biotagDM-AID were incubated with streptavidin-agarose beads and bound proteins analyzed by immunoblotting with AID antibodies (upper) or streptavidin-coupled to horseradish-peroxidase (SA-HRP, lower). (d) DM-AID binds to Sμ. Cross-linked DNA protein complexes from unstimulated or CIT-stimulated CH12BirA/biotagDM-AID cells were subjected to modified ChIP in which steptavidin-agarose replaced antibodies used in conventional ChIP. Three-fold dilutions of DNA bound to streptavidin agarose were analyzed by PCR for the presence of Sμ or the μ promoter. (e-f) Whole cell extracts derived from anti-CD40+IL-4-stimulated wild-type or AID-deficient mouse splenic B cells were immunoprecipitated with AID (e) or PTBP2 (f) antibodies and the immunoprecipitates were probed with anti-PTBP2 or anti-AID, respectively on immunoblots. E1 and E2 are two elutions of bound proteins. The data are representative of two independent experiments.