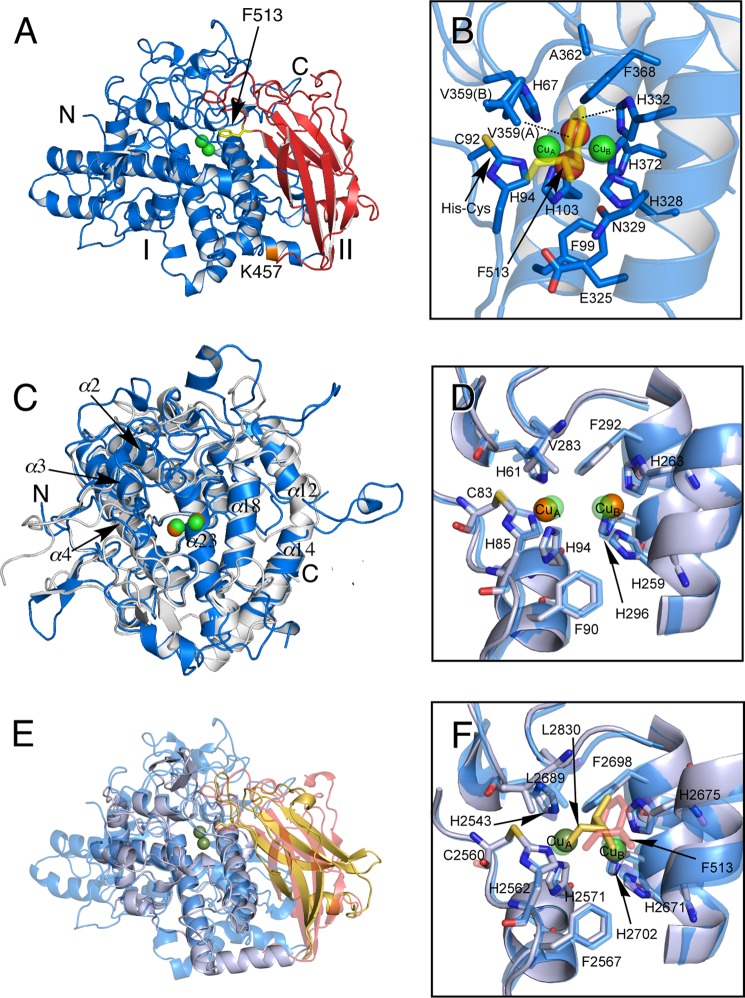
FIGURE 6.
Crystal structure of protomer of melB holo-pro-tyrosinase. A, structure of the chain A molecule. The copper-binding domain (domain I, Ser1–Phe463) is colored in blue and the C-terminal domain (domain II, Gly464–Ala616) in red. Putative proteolytic site (Lys457) is labeled and shown in orange. The N and C termini are indicated as N and C, respectively. The black arrow indicates Phe513 from the C-terminal domain covering the active site. B, active site structure viewed from the C-terminal domain. Phe513 is shown as a stick model colored in yellow. Dotted lines indicate closest contacts between atoms. C, superimposed structure of the copper-binding domain (blue) versus mushroom tyrosinase (PDB code 2Y9W) (gray). D, superimposed structure of active site structure of melB (cyan) versus mushroom tyrosinase (PDB code 2Y9W) (gray). E, superimposed structure of whole structure of melB (the copper-binding domain in cyan, and the C-terminal domain in red) versus Octopus hemocyanin (PDB code 1JS8) (gray and yellow). F, superimposed structure of active site structure of melB (cyan) versus Octopus hemocyanin (PDB code 1JS8) (gray). Both green and brown spheres and red spheres indicate copper and water, respectively. Residues are shown as sticks.