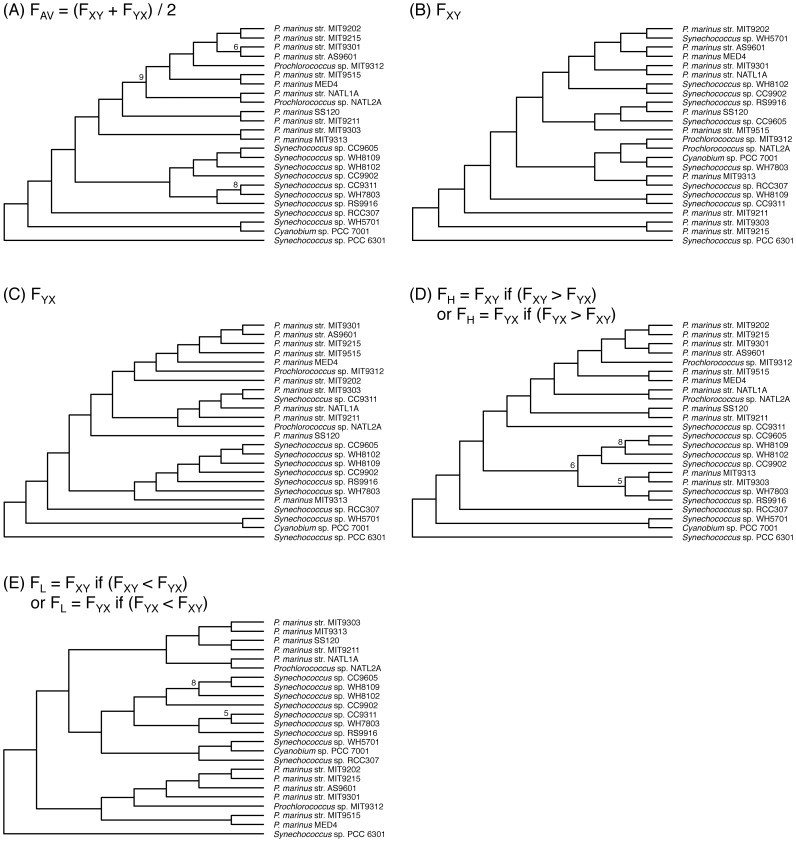
Figure 3. Phylogenetic tree constructed using a reduced gene number for the Synechococcus sp. CC9311 genome.
Ten independent databases of Synechococcus sp. CC9311 were artificially formed with 289 randomly selected genes (10% of the total gene number). Ten independent phylogenetic trees using five Prochlorococcus species and four Synechococcus species containing artificially formed Synechococcus sp. CC9311 databases were constructed using each of the distance indices used to generate Figs. 2B to 2F. A consensus tree for the ten independent trees was generated with the use of the CONSENSE program for each of the five distance indices, FAv, FXY, FYX, FH, and FL. Numbers on the branch points represent the number of identical branching patterns in ten independent trees. Branching points without numbers indicate that the number of identical branching patterns is ten. S. elongatus PCC 6301 was used as an out-group.