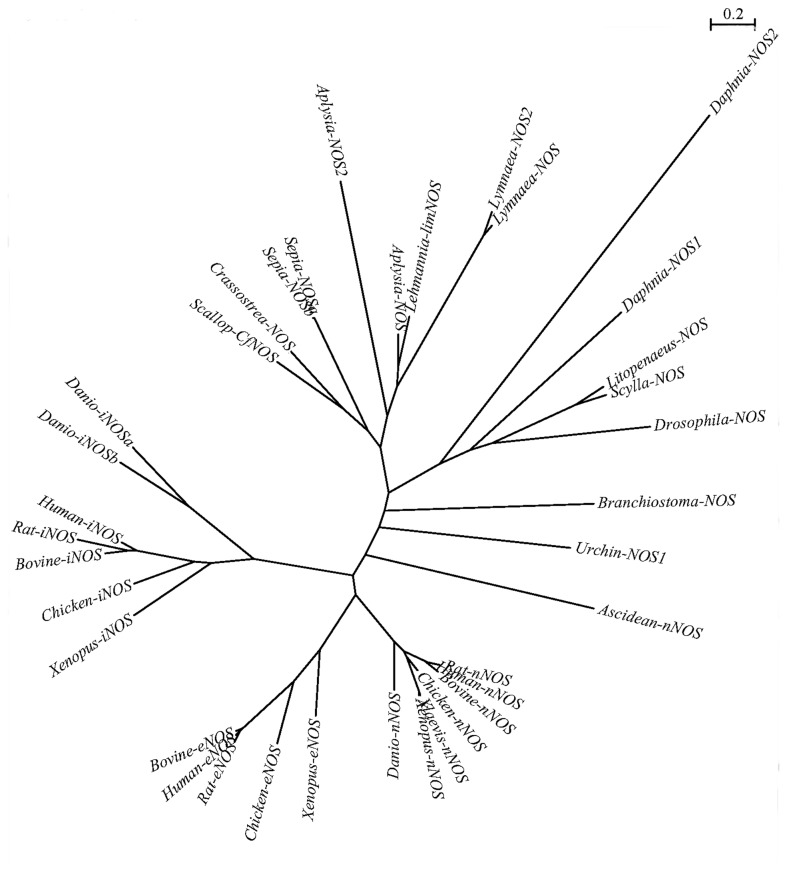
Figure 2. Maximum likelihood (ML) algorithm tree based on the amino acid sequences of NOSs.
The scale bar represents conversion of branch length to genetic distance between clades (0.1 = 10% genetic distance). The protein sequences used for phylogenetic analysis include: nNOSs from H. sapiens (AAA62405.1), Rattus norvegicus (AAC52782.1), Bos taurus (XP_002694631.2), Gallus gallus (XP_425296.2), Xenopus (Silurana) tropicalis (XP_002938130.1), Xenopus laevis (NP_001079155.1), Ciona intestinalis (XP_002120267.1), iNOSs from H. sapiens (NP_000616.3), R. norvegicus (AAC13747.1), B. Taurus (NP_001070267.1), G. gallus (NP_990292.1), X . tropicalis (XP_002935342.1), Danio rerio (NP_571735.1 and NP_001098407.1), eNOSs from H. sapiens (NP_000594.2), R. norvegicus (NP_068610.1), B. Taurus (NP_851380.2), G. gallus (AFD20677.1), X . tropicalis (ACV74251.1), and NOSs from Branchiostoma floridae (AAQ02989.1), D. melanogaster (AAC46882.1), Daphnia magna (ACQ55298.1 and ACQ55299.1), Scyllaparamamosain (CCC18661.1), Litopenaeusvannamei (ADD63793.1 and BAF73722.1), Lymnaea stagnalis (AAC17487.1 and AAW88577.1), A . californica (AAK83069.1 and AAK92211.3), Sepia officinalis (AAS93626.1 and AAS93627.1), Crassostrea gigas (EKC33784.1), S. purpuratus (XP_003729305.1).