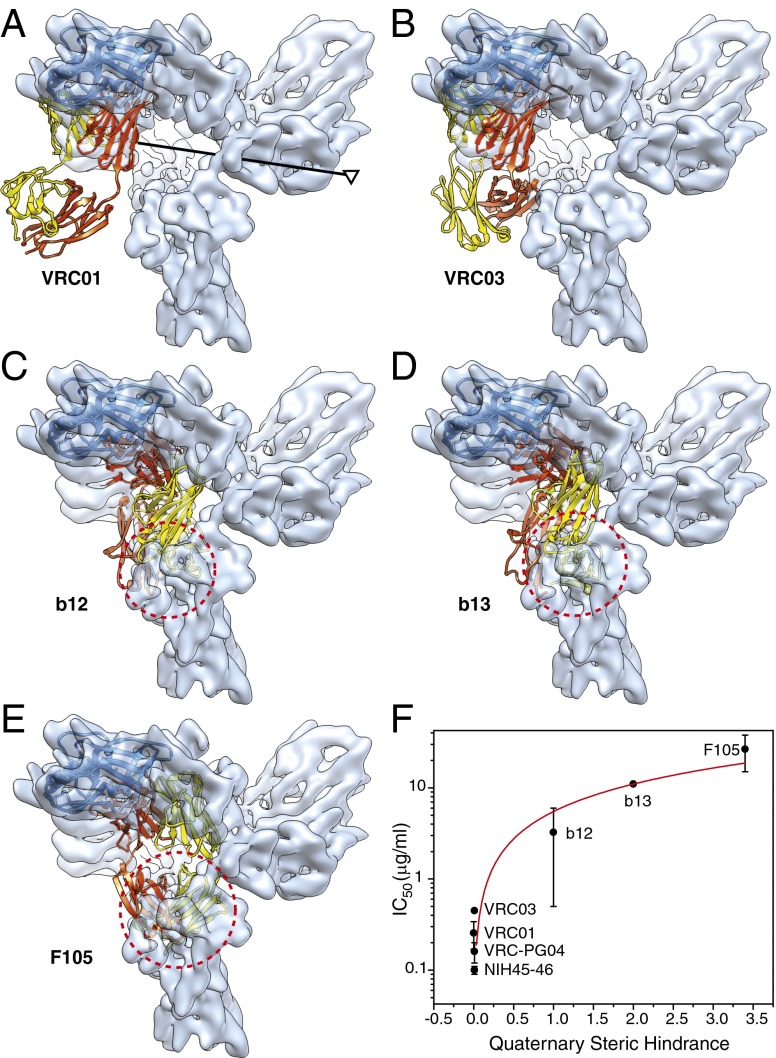
Fig. 6.
Effect of quaternary structure on virus neutralization by CD4BS antibodies. (A–E) Crystal structures of the gp120 core in complex with the CD4BS antibodies, VRC01 (A), VRC03 (B), b12 (C), b13 (D), and F105 (E) were superposed on the unliganded Env precursor map, with the Env trimer axis shown in A. The conformationally rigid gp120 outer domain was fitted to the Env trimer density map, and the outer domains in the crystal structures were aligned with the fitted outer domain (represented as a blue ribbon). The heavy and light chains of the CD4BS antibodies are colored red and yellow, respectively. The dashed red circle in C–E marks the approximate diameter of the space in the neighboring gp120 subunit that is overlapped by the antibodies, highlighting the quaternary steric hindrance that the antibodies experience when approaching their binding site on the gp120 outer domain. (F) Using the volume of the quaternary clash in the case of b12 as a normalization metric, the degree of quaternary steric hindrance encountered by each antibody as it accesses the Env trimer was quantified and was plotted against the geometric mean IC50 of the neutralization of multiple HIV-1 strains by the antibody (29–31, 43–45). The Protein Data Bank IDs of the antibodies in complex with gp120 core structures are 3NGB (A), 3SE8 (B), 2NY7 (C), 3IDX (D), and 3HI1 (E). The red line shows a linear regression of the relationship (rS: 0.9985, slope: 5.5, SE: 0.082).