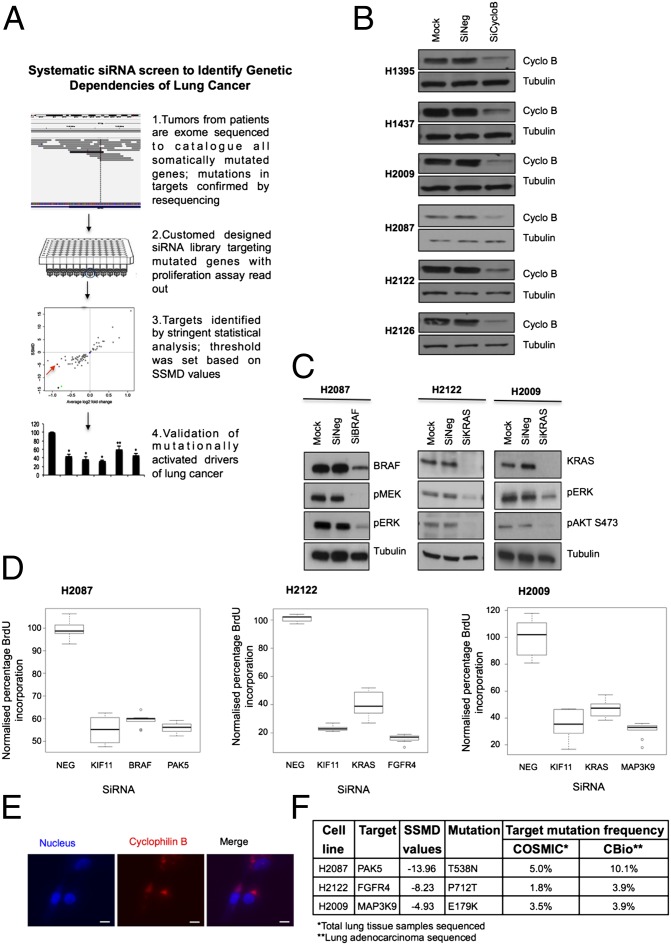
Fig. 1.
Genetic dependency screen identifies three potential genetic drivers of lung cancer. (A) Schematic of siRNA targeted screen carried out on a panel of six NSCLC cell lines. (B) Transfection efficiency for each cell line was optimized by using 100 nM siRNA against cyclophilin B (SiCycloB), 100 nM of nontargeting siRNA (SiNeg), and transfection reagent alone (Mock). (C) Cell line-specific positive controls BRAF and KRAS were knocked down by using 100 nM siRNA in respective NSCLC cells. (D) Box-and-whisker plots of statistically validated targets identified from BrdU screens, with data normalized to nontargeting siRNA (NEG), plotted with KIF11 as assay positive control, and BRAF (H2087) and KRAS (H2122, H2009) as cell line-specific positive control. (E) DY-547-cyclophilin B siRNA was used as an indicator of efficient transfection. Cell nuclei were stained with Hoechst 33342. Images captured for H2009 cells are illustrated as an example. (Scale bars: 10 µm.) (F) Validated targets identified from targeted screen based on SSMD values in three NSCLC cell lines and the frequency of their mutations following sequencing of lung cancer cells. Data compiled from COSMIC and cBio Cancer Genomics Portal databases.