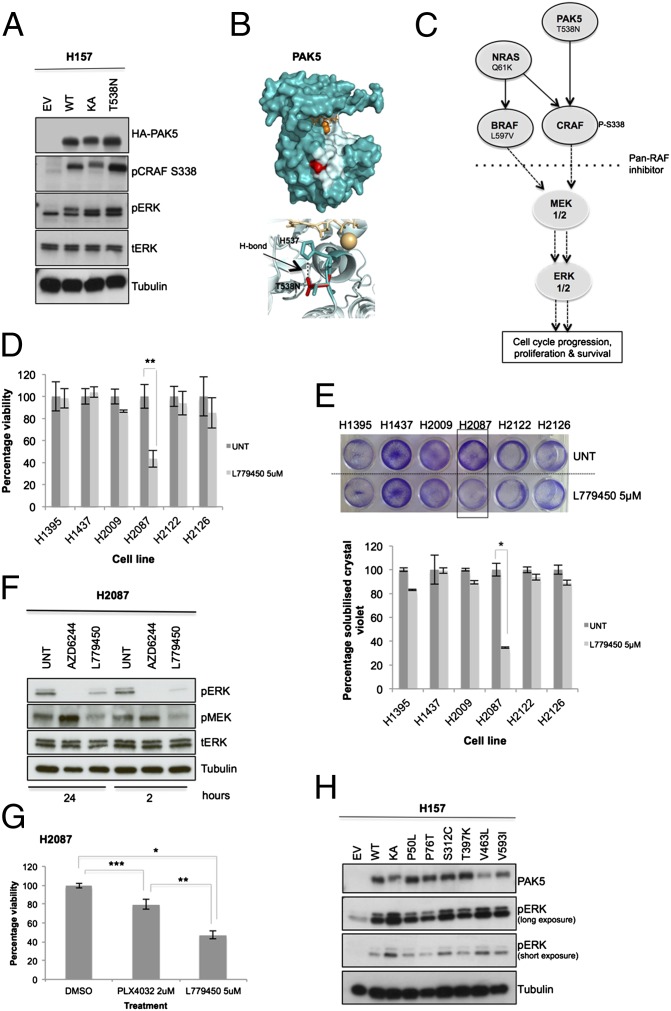
Fig. 3.
Characterization of PAK5 kinase as an actionable mutated target. (A) Empty vector (EV), WT PAK5, KA S573N, and T538N mutant were overexpressed in H157 cells, and lysates were analyzed by Western blotting. (B, Upper) Crystal structure of PAK5 kinase domain (Protein Data Bank ID code 2F57) with ATP and magnesium ions shown as orange sticks and spheres, respectively. Likely substrate binding region is shown in pale blue, based on observed binding sites for other protein kinases, including PKA (PDB ID IQ24). The mutation site of T538N, shown in red, is found at the bottom of this binding region. (B, Lower; red sticks) Analysis of the structure shows that this mutation allows more hydrogen-bonding opportunities with the neighboring histidine residue (H537; blue sticks) compared with the WT construct. (C) Mutationally activated PAK5, NRAS, and BRAF hyperactivate the MEK/ERK pathway to drive proproliferative signals and promote lung cancer cell survival. (D) The panel of six NSCLC cell lines was treated with 5 µM of the pan-RAF inhibitor L779450 for 72 h, and cytotoxicity was determined by MTT assay. Data (n = 9) were normalized to untreated cells (UNT; DMSO, <0.01%). (E, Upper) Cytotoxicity was also confirmed by colony formation assay with crystal violet staining and (E, Lower) quantified by solubilization (n = 6). (F) H2087 cells were treated with AZD6244 (3 µM) or L779450 (5 µM) for indicated time points. Inhibition of downstream substrates pMEK and pERK was analyzed. (G) H2087 cells were treated with BRAF-specific inhibitor PLX4032 (2 µM) or pan-RAF inhibitor L779450 (5 µM). Cell viability was assessed by MTT (n = 9). (H) PAK5 mutants in lung cancer identified from the COSMIC database were overexpressed in H157 cells. Three mutants—S312C, V463L, and V593I—were identified to be activating at levels comparable to KA (S573N). Error bars show SD. P values were calculated by using one-tailed paired Student t test (*P < 0.005, **P < 0.01, and ***P < 0.05 vs. DMSO control).