Abstract
Purpose
miR-452 is reported to be required for neural crest stem cell differentiation during neural crest development. However, the biological role of miR-452 in gliomas remains unclear. The aim of the present study was to evaluate the effect of miR-452 on the stem-like properties and tumorigenesis of glioma cells.
Experimental Design
The expression of miR-452 was examined in glioma cells and glioma tissues using real-time PCR. The effects of miR-452 on stem-like traits and tumorigenesis were investigated in vitro and in vivo using patient-derived glioma cells and glioma cell lines. Western blotting and luciferase reporter assays were performed to examine the negative regulation of Bmi-1, LEF1 and TCF4 by miR-452. The methylation of the miR-452 promoter region was examined by bisulfite genomic sequencing PCR.
Results
miR-452 was markedly downregulated in glioma cells and clinical glioma tissues. miR-452 levels inversely correlated with WHO grades and patient survival. miR-452 directly targeted and suppressed multiple stemness regulators, including Bmi-1, LEF1 and TCF4, resulting in reduced stem-like traits and tumorigenesis of glioma cells in vitro and in vivo. Furthermore, we demonstrated that downregulation of miR-452 in gliomas was caused by hypermethylation of its promoter region.
Conclusions
Downregulation of miR-452 plays an important role in promoting the stem-like traits and tumorigenesis of gliomas and may represent a novel prognostic biomarker and therapeutic target for the disease.
Keywords: miR-452, Glioma, Stem-like traits, Prognosis, Methylation
Introduction
Gliomas, one of the most lethal malignant tumors of the central nervous system, have been classified into four grades (I, II, III and IV) as a means of reflecting their anticipated biological behavior (1). More than half of all gliomas are glioblastoma multiforme (GBM; grade IV astrocytoma) (2). Despite modern surgical and medical treatments, the prognosis of gliomas remains very poor, with the median survival time for GBM patients being only 14.6 months (3). GBM shows a significantly similar gene-expression signature and cell phenotype to those of embryonic stem cells, whereas other glioma types show a lower degree of similarity corresponding to their lower grades (4). This suggests that molecular regulators of stem cell function might be implicated in cancer aggressiveness and poor patient survival. Therefore, understanding the precise molecular mechanisms underlying the stem cell-like properties of cancer cells could provide new insights into the pathogenesis of gliomas and lead to more effective anticancer therapy strategies.
The molecular pathways controlling the function of stem cells, such as the Bmi-1 and Wnt/β-catenin pathways, have been demonstrated to play important roles in tumor progression (5, 6). For instance, Bmi-1 is essential for the “stemness” of both normal stem cells and cancer stem cells, such as leukemias, gliomas and hepatocellular carcinomas (7–10). Abdouh et al. reported that Bmi-1 maintains glioma stem-like cells in an undifferentiated state, and that knockdown of Bmi-1 induces glioma cell differentiation and prevents tumor formation in xenografts (9). Similarly, the Wnt/β-catenin signaling pathway has been shown to be important for the regulation of the stemness properties and tumorigenicity of glioma cells (11). Silencing β-catenin in FoxM1-overexpressing glioma cells diminishes neurosphere formation and tumor initiation, indicating that Wnt/β-catenin signaling is required for the FoxM1-mediated stem-like phenotype of gliomas (12).
MicroRNAs (miRNAs) are small, noncoding RNAs that have recently emerged as important molecules involved in post-transcriptional gene regulation (13). Multiple miRNAs have been reported to play critical roles in the stem-like traits and aggressiveness of cancers (14–16). Overexpression of miR-124 can reduce neurosphere formation, CD133+ subpopulation and glioma cell tumorigenicity and invasion (16). Interestingly, the miRNA-expression profiles of gliomas are similar to those of neural precursor cells (17), suggesting that the miRNAs involved in stemness regulation may also contribute to glioma pathogenesis.
It has been reported that miR-452 is required for the differentiation of neural crest stem cell-derived tissues, and miR-452 knockdown during neural crest development leads to craniofacial defects (18). Herein, we report that miR-452 is downregulated in human gliomas, especially, high-grade, undifferentiated gliomas. We find that upregulation of miR-452 suppresses glioma stem-like traits and tumorigenesis both in vitro and in vivo, through the inhibition of multiple stemness regulators, including Bmi-1, LEF1 and TCF4. These results uncover a novel regulatory mechanism underlying the stem-like properties of glioma cells.
Materials and Methods
Cell lines and primary cultured tumor cells
Primary normal human astrocytes (NHA) were purchased from ScienCell Research Laboratories (Carlsbad, CA) and cultured according to the manufacturer’s instruction. Fresh brain tumor tissues obtained from the First Affiliated Hospital of Sun Yat-sen University were collected and processed within 30 min after resection. Primary cultured tumor cells were obtained after mechanical dissociation as previously described (19). Glioma cell lines U87MG, A172, U251MG, LN-340, LN-464, LN-428, LN-18, U118MG, U138MG, LN-443, D247MG and LN-229 were provided by Dr. Shi-Yuan Cheng’s laboratory at Northwestern University, Chicago, IL, and cultured in DME medium supplemented with 10% fetal bovine serum (HyClone, Logan, UT). All cell lines were authenticated by short tandem repeat fingerprinting at IDEXX RADIL (Columbia, MO, USA) or Services at SYSU Forensic Medicine Lab (Guangzhou, China).
Plasmids, virus production and infection of target cells
The human miR-452 precursor with 500-bp genomic flanking sequences on each side was amplified by PCR from genomic DNA and cloned into a retroviral pMSCV-puro vector. Human Bmi-1, LEF1 and TCF4 open reading frames were amplified by PCR and were cloned into the pMSCV-neo vector (Clontech, Palo Alto, CA). The 3′untranslated regions (3′UTRs) of the human Bmi-1, LEF1 and TCF4 genes, generated by PCR amplification from NHA, were cloned into a modified pGL3 luciferase reporter plasmid (Promega, Madison, WI). The point mutations in the tentative miR-452-binding seed regions in the 3′UTRs of the Bmi-1, LEF1 and TCF4 genes were created using the QuikChange Mutagenesis kit (Stratagene, La Jolla, CA). pTOP-Flash and pFOP-Flash plasmids were used to determine β-catenin transcriptional activity. miR-452 mimics and negative control oligonucleotides were purchased from Ribo Biotech (Guangzhou, China). Transfection of plasmids or oligonucleotides was performed using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instruction. Stable cell lines expressing miR-452 and vector were generated via retroviral infection using HEK293T cells, as previously described, and selected with 0.5 μg/ml puromycin for 10 days (20). The primers used for plasmid construction are listed in Supplementary Materials and Methods.
Tissue specimens and patient information
A total of 88 paraffin-embedded, archived clinical glioma specimens, including WHO grade I–IV tumors and freshly snap-frozen glioma tissues were histopathologically diagnosed at the First Affiliated Hospital of Sun Yat-sen University from 2000 to 2010. The clinical information related to the samples has been summarized in Supplementary Table S1. Normal brain tissues were obtained from individuals who died in traffic accidents and confirmed to be free of any pre-existing pathologically detectable conditions. For the use of these clinical materials for research purposes, patients’ consent and approval from the Institutional Research Ethics Committee were obtained.
Western blotting (WB) analysis
WB was performed as previously described (20) using anti-Bmi-1 (Millipore, Billerica, MA); anti-TCF4 and anti-LEF1 (Cell Signaling, Danvers, MA) antibodies. The blotting membranes were stripped and re-probed with an anti-α-tubulin antibody (Sigma, Saint Louis, MO).
Intracranial brain tumor xenografts, IHC and H&E staining
Indicated glioma cells were stereotactically implanted into the brains of individual mice (n = 5/group). One group of mice was monitored daily and euthanized when the first mice were moribund. For Kaplan–Meier analysis, another group of mice was monitored daily and euthanized when the mice were moribund. Whole brains were removed, paraffin-embedded, sectioned at 4-μm intervals and then stained with H&E or with anti-Nestin (Chemicon, Billerica, MA) and anti-GFAP (Dako, Glostrup, Denmark) antibodies. Images were captured using the AxioVision Rel.4.6 computerized image analysis system (Carl Zeiss).
Bisulfite genomic sequencing
Genomic DNA extracted from NHA, glioma cell lines and clinical samples was treated with bisulfite using the Epitect Bisulfite kit (Qiagen, Valencia, CA) according to the manufacturer’s instruction. The bisulfite-treated DNA was amplified with bisulfite-sequencing PCR primers designed by MethPrimer. Analysis for the methylation status of the GABRE promoter region was based on the following primers: forward, 5′-GAGGAGTGTTTGTTATTAGGTTAT-3′ and reverse, 5′-TAAACATCTTA TTAATCCTCCAATC-3′. The PCR products were cloned with the pGEM-T Easy Vector System (Promega). A single clone each from the NHA, glioma cell lines and clinical samples was selected and sequenced.
Statistical analysis
All statistical analyses were carried out using the SPSS 10.0 statistical software package. The chi-square test was used to analyze the relationship between miR-452 expression and the clinicopathological characteristics. Bivariate correlations between study variables were calculated by Spearman’s rank correlation coefficients. Survival curves were plotted by the Kaplan-Meier method and compared by the log-rank test. The significance of various variables for survival was analyzed by univariate and multivariate Cox regression analyses. P <0.05 in all the experiments was considered statistically significant.
Results
miR-452 downregulation in gliomas correlates with improved patient prognosis
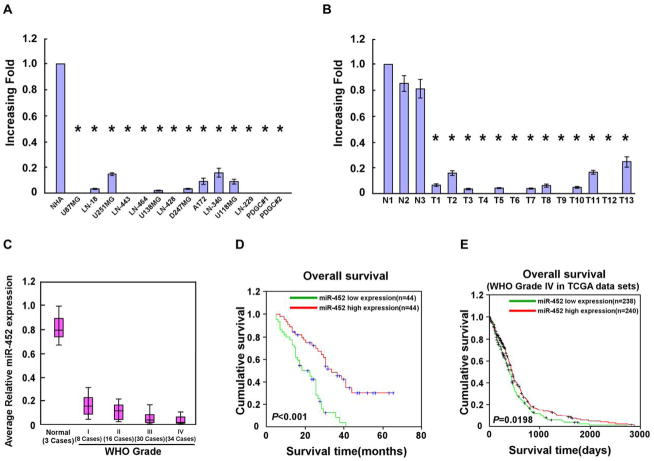
To investigate the biological role of miR-452 in the stem-like traits and aggressiveness of gliomas, we first examined miR-452 expression in glioma cells and clinical glioma tissues. The expression of miR-452 was differentially reduced in all 12 glioma cell lines and 2 patient-derived glioma cells (PDGCs) compared with that in NHA, and in 13 glioma samples compared with that in normal brain tissues (n = 3), indicating that miR-452 is downregulated in gliomas (Fig. 1A and B).
Figure 1. Reduced miR-452 expression in gliomas correlates with poor prognosis.
A–B. Real-time PCR analysis of miR-452 expression in 12 indicated glioma cell lines, 2 PDGCs and NHA (A), and in 13 glioma tissues and 3 normal brain tissues (B). Transcript levels were normalized to U6 expression. C. Statistical analysis of miR-452 expression in normal brain tissues (n = 3) and glioma specimens of different WHO grades (n = 88). D. Kaplan-Meier curves of glioma patients with low- versus high-expression of miR-452 (n = 88; P < 0.001). E. Kaplan-Meier curves of glioblastoma patients with low- versus high-expression of miR-452 in TCGA data sets (n = 478; P = 0.0198). Error bars represent the mean ± SD of three independent experiments. *, P < 0.05.
The expression of miR-452 was further examined in 88 clinical glioma samples. Statistical analysis revealed that miR-452 expression inversely correlated with the WHO histological grade (P = 0.018) and patient survival (P = 0.008; Fig. 1C and Supplementary Table S2). Kaplan–Meier analysis and the log-rank test revealed that decreased miR-452 expression correlated with shorter patient survival time (P < 0.001; Fig. 1D and Supplementary Fig. S1). Importantly, univariate and multivariate analyses revealed that miR-452 expression (P < 0.001) and WHO histological grade (P < 0.001) were each recognized as an independent prognostic factor for glioma patients (Supplementary Table S3). Furthermore, through analyzing The Cancer Genome Atlas (TCGA) database, we also found that decreased miR-452 expression correlated with shorter glioblastoma patient survival time (n = 478, P = 0.0198; Fig. 1E), and that miR-452 expression (P = 0.045) was an independent prognostic factor for glioblastoma patients (Supplementary Tables S4 and S5). Taken together, these data suggest that miR-452 downregulation might represent a useful independent biomarker for the prognosis of patients with gliomas.
Upregulation of miR-452 reduces glioma cell stem-like traits
To examine the effect of miR-452 dysregulation on glioma stem-like traits, 2 PDGCs and U87MG and LN443 glioma cell lines stably overexpressing miR-452 were established (Fig. 2A and Supplementary Fig. S2A). As shown in Fig. 2B and Supplementary Fig. S2B, overexpression of miR-452 markedly reduced the proportion of side-population (SP) cells in all 4 glioma cells. However, the proportion of SP cells was increased in glioma cells transfected with a miR-452 inhibitor (Supplementary Fig. S2C). The expression of miR-452 in SP cells was showed to be lower than that in non-SP cells (data not shown). Meanwhile, miR-452 overexpression significantly reduced the mRNA-expression levels of multiple pluripotency factors, including ABCG2, KLF4, SOX2, OCT4, NANOG and c-Myc (Fig. 2C and Supplementary Fig. S2D) (21). Furthermore, a neurosphere assay showed that miR-452-transduced cells formed smaller and fewer neurospheres than vector controls (Fig. 2D and Supplementary Fig. S2E). To test whether miR-452 is involved in glioma differentiation, the neurospheres formed by miR-452- and vector-transduced cells and cultured under differentiating conditions with 5% serum. Immunofluorescence analysis revealed that overexpression of miR-452 led to decreased Nestin (a neural stem cell marker) expression and increased glial fibrillary acidic protein (GFAP; an astrocyte marker) compared with vector controls, indicating that miR-452 induces the differentiation of glioma stem-like cells (Fig. 2E and Supplementary Fig. S2F). Collectively, our results suggest that miR-452 overexpression reduces the stem-like traits of glioma cells.
Figure 2. Upregulation of miR-452 reduces glioma stem-like traits.
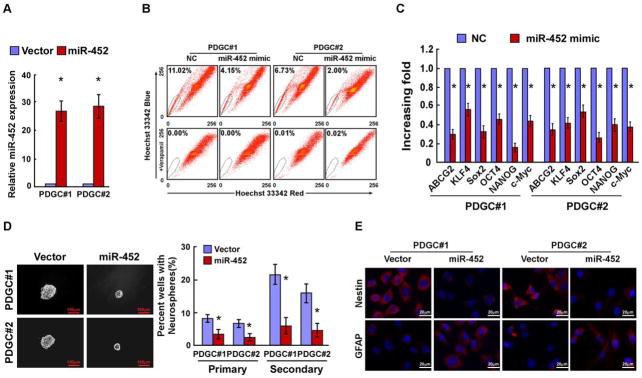
A. Real-time PCR analysis of miR-452 expression in the indicated cells. Transcript levels were normalized to U6 expression. B. Quantification of SP cells among the indicated cells. C. Relative expression of pluripotency factors in the indicated cells. Transcript levels were normalized to GAPDH expression. D. Representative micrographs (left panel) and quantification (right panel) of neurospheres formed in the indicated cells. Scale bars, 100 μm. E. Staining of Nestin and GFAP in neurosphere-derived cells grown under adherent differentiation conditions. Scale bars, 20 μm. Error bars represent the mean ± SD of three independent experiments. *, P < 0.05.
miR-452 directly downregulates multiple stemness regulators
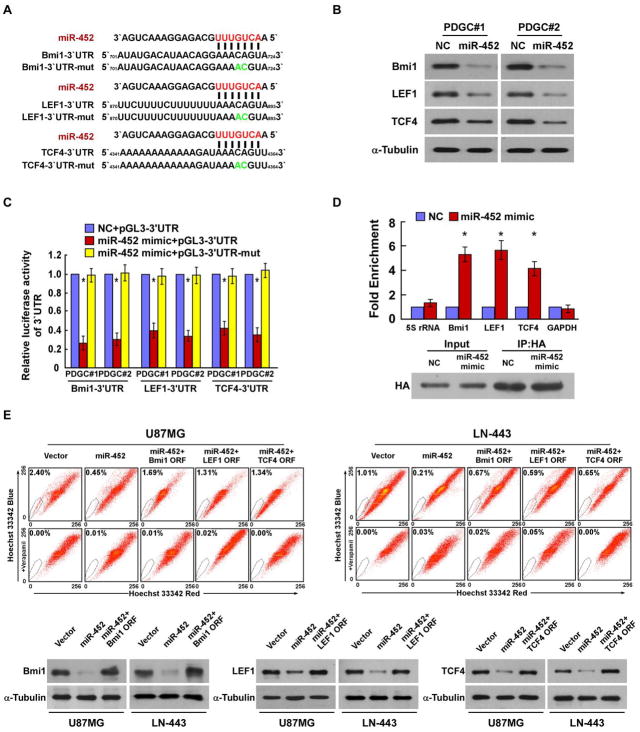
Analysis using a publicly available algorithm (TargetScan) showed that multiple key regulators of stemness, including Bmi-1, LEF1 and TCF4, might be the potential targets of miR-452 (Fig. 3A). WB analysis revealed that the expressions of Bmi-1, LEF1 and TCF4 were drastically decreased in miR-452-transduced cells (Fig. 3B and Supplementary Fig. S3A). In addition, miR-452 overexpression significantly attenuated the activities of luciferase reporters linked to the 3′UTRs of Bmi-1, LEF1 and TCF4, but failed to reduce the luciferase activities of the mutated 3′UTRs of these transcripts (Fig. 3C and Supplementary Fig. S3B). However, the inhibitory effects of miR-452 on the expressions of Bmi-1, LEF1 and TCF4 and luciferase reporter activities were abrogated by miR-452 inhibition (Supplementary Fig. S3C and D). Furthermore, miRNP IP analysis revealed that miR-452 specifically associated with the 3′UTRs of Bmi-1, LEF1 and TCF4, but not with GAPDH (Fig. 3D and Supplementary Fig. S3E), thus providing additional evidence that miR-452 directly targets the 3′UTRs of Bmi-1, LEF1 and TCF4. Importantly, restoration of Bmi-1, LEF1 or TCF4 expression partially, but significantly, rescued the proportion of SP cells in miR-452-transduced cells, indicating that Bmi-1, LEF1 and TCF4 genes are regulatory targets that contributed to miR-452 downregulation mediated-glioma stem-like traits (Fig. 3E). Moreover, we found that the luciferase activity of a TOP-Flash reporter and the expressions of SNAI1, AXIN2, MMP7, Cyclin D1 and MET, the established downstream targets of the Wnt/β-catenin pathway, were also decreased in the miR-452-transduced cells compared to control cells (Supplementary Fig. S4), suggesting that miR-452 overexpression represses Wnt/β-catenin signaling.
Figure 3. miR-452 directly downregulates multiple stemness regulators.
A. Predicted miR-452 target sequences in the Bmi-1–3′UTR, LEF1–3′UTR and TCF4–3′UTR and mutants containing two mutated nucleotides in 3′UTR of these transcripts. B. Western blotting analysis of the expressions of Bmi-1, LEF1 and TCF4 in the indicated cells. α-Tubulin was used as a loading control. C. Luciferase activities of reporters containing the 3′UTRs or mutated-3′UTRs of Bmi-1, LEF1 and TCF4 in the indicated cells transfected with negative control (NC) or miR-452 mimic. D. Upper panel: miRNP IP assay showing the association of miR-452 with Bmi-1, LEF1 and TCF4. GAPDH was used as a negative control and 5S rRNA was used as a control for overall expression levels. Lower panel: Western blotting analysis of HA-Ago1. E. Upper panel: Restoration of Bmi-1, LEF1 and TCF4 expression partially, but significantly, rescued the proportion of SP cells in miR-452-transduced cells. Lower panel: Western blotting analysis of Bmi-1, LEF1 and TCF4 protein expression in the indicated cells. α-Tubulin was used as a loading control. Error bars represent the mean ± SD of three independent experiments. * P, < 0.05.
Notably, miR-452 levels in nine freshly collected clinical glioma samples were inversely correlated with the expressions of Bmi-1 (r = −0.867; P = 0.002), LEF1 (r = −0.767; P = 0.016) and TCF4 (r = −0.817; P = 0.007), further confirming the inhibitory effects of miR-452 on Bmi-1, LEF1 and TCF4 in gliomas (Fig. 4A and B).
Figure 4. Clinical relevance of Bmi-1, LEF1 and TCF4 expression and miR-452 in human glioma samples.
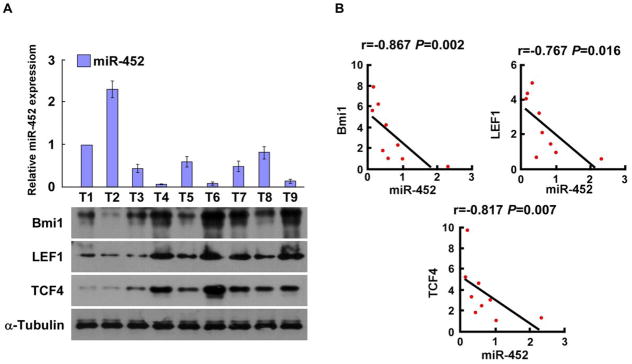
A–B. Quantification (A) and correlations (B) of miR-452 levels and the expression of Bmi-1, LEF1 and TCF4 in nine freshly collected human glioma samples. Error bars represent the mean ± SD of three independent experiments. * P, < 0.05.
Upregulation of miR-452 decreases tumorigenesis of glioma cells in vivo
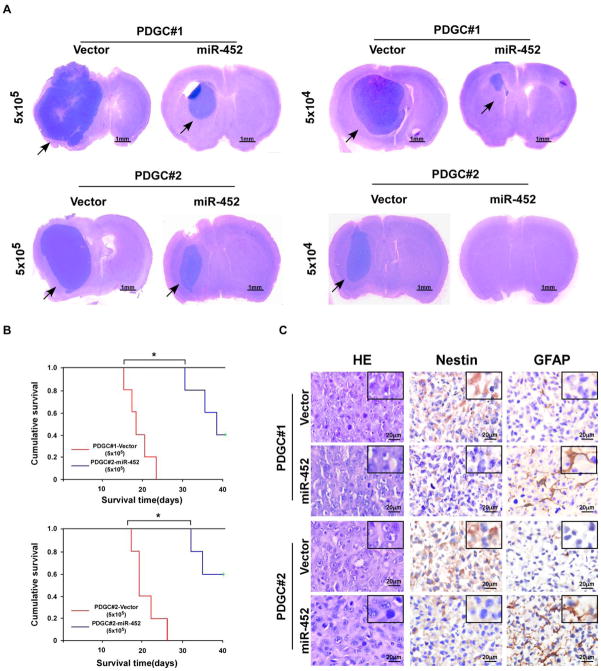
To determine the role of miR-452 in modulating glioma tumorigenesis in vivo, we stereotactically implanted miR-452- or vector-transduced PDGCs into the brains of nude mice. As shown in Fig. 5A and Supplementary Table S6, miR-452-transduced cells formed much smaller tumors and displayed lower tumorigenicity, demonstrating that miR-452 overexpression reduces PDGC tumorigenesis in vivo. Kaplan–Meier analysis revealed that mice implanted with miR-452-transduced cells survived significantly longer than control mice (Fig. 5B). IHC analysis showed that intracranial tumors formed by miR-452-transduced PDGCs had fewer Nestin-positive tumor cells and more GFAP-expressing cells than the control tumors (Fig. 5C). Consistently, these results suggest that miR-452 overexpression also inhibit the tumorigenicity of U87MG glioma cells in vivo (Supplementary Fig. S5A–C).
Figure 5. miR-452 overexpression reduces glioma cell tumorigenesis in vivo.
A. Representative images of tumor formed by miR-452- and vector-transduced cells in the mouse brai[a–z].[a–z]iR-452- and vector-transduced cells were stereotactically implanted into the brains (n = 5/group). Scale bars, 1 mm. B. Kaplan–Meier survival curves of mice inoculated with the indicated cells. C. H&E and IHC staining for Nestin and GFAP in tumors formed by miR-452- and vector-transduced cells. Scale bars, 20 μm.
miR-452 is downregulated by promoter hypermethylation
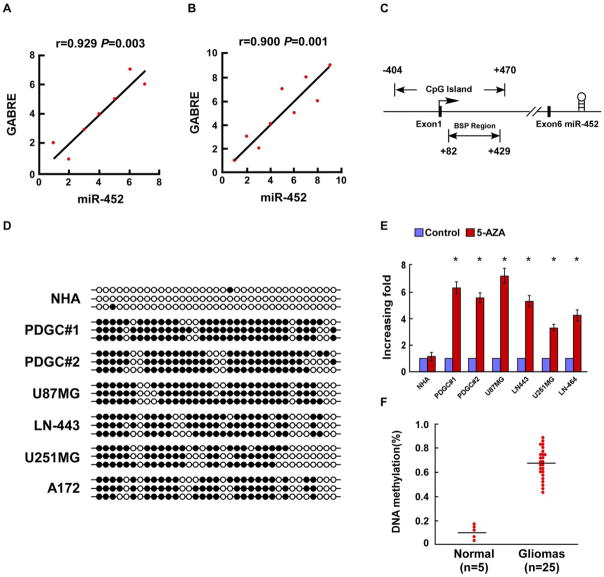
The miR-452 coding sequence is located in the intron of the GABRE gene at Xq28. In agreement with a previous study (22), we found that miR-452 expression correlated with GABRE mRNA expression in glioma cell lines (P = 0.003) and glioma tissues (P = 0.001; Fig. 6A and B), suggesting that miR-452 is likely to be co-transcribed with GABRE mRNA. Promoter methylation has been reported to control GABRE expression in gastric cancer and uterine leiomyoma (23, 24). Therefore, we investigated whether the reduced miR-452 in gliomas was a result of promoter hypermethylation. Analysis of the GABRE promoter region using the UCSC genome browser (http://genome.ucsc.edu/) indicated a CpG island located between −404 bp and +470 bp, relative to the transcription start site (Fig. 6C). Furthermore, bisulfite genomic sequencing PCR (BSP) showed that the CpG island within the GABRE promoter was hypermethylated in glioma cell lines and PDGCs, but not in NHA (Fig. 6D). Moreover, we found that miR-452 expression was significantly increased in glioma cell lines and PDGCs treatment with the methylase inhibitor 5-aza-dC, but not in NHA (Fig. 6E). Consistent with the data from cultured glioma cells, the CpG island in the GABRE promoter was also found to be hypermethylated in clinical glioma samples (n = 25), but not in normal brain tissue (n = 5; Fig. 6F). These results indicate that the reduced miR-452 expression in gliomas is attributable to promoter hypermethylation.
Figure 6. Downregulation of miR-452 in gliomas is due to promoter hypermethylation.
A. The expression of miR-452 correlated with GABRE mRNA expression in seven glioma cell lines. B. The expression of miR-452 correlated with GABRE mRNA expression in nine glioma tissue samples. C. Schematic illustration of the position of the miR-452 stem loop within the GABRE genomic sequence. D. Bisulfite sequencing analysis of NHA, 4 indicated glioma cell lines and 2 PDGCs. Three clones of PCR products from each sample of bisulfite-treated DNA were sequenced. Filled and open circles indicate methylation and nonmethylation, respectively. E. miR-452 was upregulated in the indicated glioma cell lines following 5-aza-dC treatment for 72 h. F. Percentage DNA methylation in GABRE promoter CpG islands based on the bisulfite sequencing results. Hypermethylation was observed in glioma samples (n = 25). Error bars represent the mean ± SD of three independent experiments. *, P < 0.05.
Discussion
This study establishes an important role of miR-452 in the inhibition of glioma tumorigenesis. We find that overexpression of miR-452 directly suppresses the expressions of Bmi-1, LEF1 and TCF4 in glioma cells, attenuates glioma stem-like phenotypes in vitro and inhibits glioma tumorigenesis in the brains of animals. Conversely, inhibition of miR-452 induces expression of the above stemness regulators. Furthermore, we demonstrate that the expression levels of miR-452 are decreased in clinical glioma samples via promoter hypermethylation. Taken together, our results reveal a novel mechanism by which miR-452 regulates glioma stem-like phenotypes through modulating multiple stemness regulators, suggesting that miR-452 might function as a tumor-suppressive miRNA in human gliomas.
A better understanding of the critical pathways involved in stem-like traits is important for identifying new molecular targets for eradicating gliomas. Bmi-1, a component of the polycomb repressor complex, is reported to be essential for the proliferation and self-renewal of leukemic and hematopoietic stem-like cells through binding to trimethylated H3K27 in the CDKN2A promoter and repressing the expression of p16INK4A and p19ARF (10, 25, 26). However, it has been also reported that Bmi-1 is highly expressed in gliomas and promotes glioma stem-like cell proliferation and undifferentiation in a p16INK4A/p19ARF- independent manner (27, 28). Furthermore, the constitutive activation of the Wnt/β-catenin pathway has been shown to lead to reprogramming and generating a stem-like phenotype in gliomas through the activation of several downstream genes, including c-Myc, NANOG, OCT4 and SOX2 (11, 12, 29–31). As both the Bmi-1 and Wnt/β-catenin pathways are critical regulators of stemness, thus inhibition of these two pathways would significantly reduce the stem-like properties of gliomas. Herein, we found that miR-452 could directly target and suppress the expressions of Bmi-1, LEF1 and TCF4, and thereby reduce neurosphere formation and decrease the proportion of SP cells. Therefore, our data reveals a novel role of miR-452 downregulation in glioma tumorigenesis through the modulation of glioma stem-like traits. Previously, Fang et al. have reported that miR-452 could be downregulated by the stem cell transcription factor SOX2 in GBM (32). In the current study, we found that the expression levels of multiple stem cell factors, including SOX2, were significantly decreased in miR-452-overexpressing glioma cells. Meanwhile, we showed that miR-452 was markedly reduced in gliomas and that the downregulation of miR-452 in gliomas cells was caused by hypermethylation of its promoter region. Thus, integrating previous study and our results suggest that there may be a feedback loop in the expressions of miR-452 and SOX2.
Epigenetic modifications, such as DNA promoter methylation, have been demonstrated to play crucial roles in the regulation of gene expression in various physiological and pathological processes (33). During development and carcinogenesis, hypermethylation of tumor-suppressor genes and hypomethylation of the CpG islands in oncogenes regulate a number of vital cellular processes. Aberrant site-specific DNA methylation is therefore a potentially useful biomarker for detecting cancer and predicting patient survival (34). Emerging evidence also strongly suggests that DNA methylation could lead to dysregulation of miRNAs in cancer (35). The role of miR-452 during development has been established. miR-452 is essential for the proper development of neural crest cell-derived tissues through regulating an epithelial-mesenchymal signaling cascade by directly targeting Wnt5a in the neural crest cell-derived mesenchyme (18). Moreover, treatment of mouse embryos with the demethylating agent 5-aza-dC resulted in inhibition of development from morula to blastocyst and elevation of miR-452 (36). These data not only suggest that miR-452 plays a role in cell differentiation during development but also indicate that miR-452 expression is tightly regulated through the methylation of its gene. Consistent with these studies, we showed that treatment of PDGCs and glioma cells with 5-aza-dC significantly increased miR-452 expression, and the CpG island in the miR-452 promoter was hypermethylated in glioma cells and clinical glioma samples. Importantly, reduced miR-452 levels significantly correlated with the WHO grades and overall survival of glioma patients. Thus, our data strongly suggest that miR-452 promoter methylation could be a potential predictor of glioma progression and patient survival.
It has been reported that miR-452 is downregulated in human breast cancer (37). Our result is consistent with this observation. We found that miR-452 was significantly downregulated in glioma cell lines and human primary glioma tissues when compared with that in NHA and normal brain tissues. However, miR-452 has been found to be upregulated in hepatocellular carcinomas (38) and lymph node-positive urothelial carcinomas (39), suggesting that miR-452 could have a tumor-promoting role in distinct types of human cancers. This signifies an important issue that the biological activities of miRNAs could be determined by the distinct context in various microenvironments. Indeed, accumulating evidence demonstrates that an individual miRNA could have diverse functions under different cellular contexts, likely depending on the availability of specific targets or downstream effectors (40, 41). Nonetheless, our results reveal an important role of miR-452 in inhibiting glioma tumorigenesis, thereby providing new mechanistic insights that could establish miR-452 as a novel and potentially useful biomarker for the prognosis of patients with gliomas. Recently, Kim et al. selected 121 miRNAs, including miR-452, to identify clinically and genetically distinct glioblastoma subclasses, and they found that miR-452 had relatively high expression in the “neuro-mesenchymal precursors” subset but had relatively low expression in the “neural”, “oligoneural”, “astrocytic” and “radial glial” subsets (42). Thus, it is worthy to further investigate the effect of miR-452 in organizing and maintaining the phenotypic and molecular architecture of glioblastoma subsets.
Translational Relevance.
The molecular regulators of stem cell stemness are implicated in cancer aggressiveness and poor patient survival. Herein, we found that miR-452, a microRNA required for neural crest stem cell differentiation during neural crest development, is markedly downregulated in gliomas, and its expression levels inversely correlate with WHO grades and patient survival. miR-452 directly targeted and suppressed multiple stemness regulators, including Bmi-1, LEF1 and TCF4, resulting in reduced stem-like traits and tumorigenesis of glioma cells in vitro and in vivo. Furthermore, we demonstrated that downregulation of miR-452 in gliomas is caused by hypermethylation of its promoter region. Taken together, our results suggest that miR-452 downregulation plays an important role in promoting the stem-like traits and tumorigenesis of gliomas and may represent a novel prognostic biomarker and therapeutic target for the disease.
Acknowledgments
Financial support: Supported by Natural Science Foundation of China (No. 81071780, 81272198, U1201121, 30900569); The Science and Technology Department of Guangdong Province (No. S2011020002757, S2012040007113); China Postdoctoral Science Foundation (No.2011M501366); Ministry of Education of China (20100171110080).
Footnotes
Competing interests: No potential conflicts of interest were disclosed.
References
- 1.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Brien CA, Kreso A, Jamieson CH. Cancer stem cells and self-renewal. Clin Cancer Res. 2010;16:3113–20. doi: 10.1158/1078-0432.CCR-09-2824. [DOI] [PubMed] [Google Scholar]
- 6.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 7.Yu CC, Lo WL, Chen YW, Huang PI, Hsu HS, Tseng LM, et al. Bmi-1 Regulates Snail Expression and Promotes Metastasis Ability in Head and Neck Squamous Cancer-Derived ALDH1 Positive Cells. J Oncol. 2011;2011 doi: 10.1155/2011/609259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiba T, Seki A, Aoki R, Ichikawa H, Negishi M, Miyagi S, et al. Bmi1 promotes hepatic stem cell expansion and tumorigenicity in both Ink4a/Arf-dependent and -independent manners in mice. Hepatology. 2010;52:1111–23. doi: 10.1002/hep.23793. [DOI] [PubMed] [Google Scholar]
- 9.Abdouh M, Facchino S, Chatoo W, Balasingam V, Ferreira J, Bernier G. BMI1 sustains human glioblastoma multiforme stem cell renewal. J Neurosci. 2009;29:8884–96. doi: 10.1523/JNEUROSCI.0968-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–60. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 11.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–50. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 12.Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, et al. FoxM1 promotes beta-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell. 2011;20:427–42. doi: 10.1016/j.ccr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Park DM, Rich JN. Biology of glioma cancer stem cells. Mol Cells. 2009;28:7–12. doi: 10.1007/s10059-009-0111-2. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Dutta A, Abounader R. The role of microRNAs in glioma initiation and progression. Front Biosci. 2012;17:700–12. doi: 10.2741/3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia H, Cheung WK, Ng SS, Jiang X, Jiang S, Sze J, et al. Loss of brain-enriched miR-124 microRNA enhances stem-like traits and invasiveness of glioma cells. J Biol Chem. 2012;287:9962–71. doi: 10.1074/jbc.M111.332627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavon I, Zrihan D, Granit A, Einstein O, Fainstein N, Cohen MA, et al. Gliomas display a microRNA expression profile reminiscent of neural precursor cells. Neuro Oncol. 2010;12:422–33. doi: 10.1093/neuonc/nop061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheehy NT, Cordes KR, White MP, Ivey KN, Srivastava D. The neural crest-enriched microRNA miR-452 regulates epithelial-mesenchymal signaling in the first pharyngeal arch. Development. 2010;137:4307–16. doi: 10.1242/dev.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruna A, Darken RS, Rojo F, Ocana A, Penuelas S, Arias A, et al. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11:147–60. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Zhang N, Song LB, Liao WT, Jiang LL, Gong LY, et al. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res. 2008;14:3319–26. doi: 10.1158/1078-0432.CCR-07-4054. [DOI] [PubMed] [Google Scholar]
- 21.Kashyap V, Rezende NC, Scotland KB, Shaffer SM, Persson JL, Gudas LJ, et al. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009;18:1093–108. doi: 10.1089/scd.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Lee CG. Role of miR-224 in hepatocellular carcinoma: a tool for possible therapeutic intervention? Epigenomics. 2011;3:235–43. doi: 10.2217/epi.11.5. [DOI] [PubMed] [Google Scholar]
- 23.Zhao J, Liang Q, Cheung KF, Kang W, Lung RW, Tong JH, et al. Genome-wide identification of Epstein-Barr virus-driven promoter methylation profiles of human genes in gastric cancer cells. Cancer. 2012 doi: 10.1002/cncr.27724. [DOI] [PubMed] [Google Scholar]
- 24.Maekawa R, Yagi S, Ohgane J, Yamagata Y, Asada H, Tamura I, et al. Disease-dependent differently methylated regions (D-DMRs) of DNA are enriched on the X chromosome in uterine leiomyoma. J Reprod Dev. 2011;57:604–12. doi: 10.1262/jrd.11-035a. [DOI] [PubMed] [Google Scholar]
- 25.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–7. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–5. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Gong LY, Song LB, Jiang LL, Liu LP, Wu J, et al. Oncoprotein Bmi-1 renders apoptotic resistance to glioma cells through activation of the IKK-nuclear factor-kappaB Pathway. Am J Pathol. 2010;176:699–709. doi: 10.2353/ajpath.2010.090502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruggeman SW, Hulsman D, Tanger E, Buckle T, Blom M, Zevenhoven J, et al. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell. 2007;12:328–41. doi: 10.1016/j.ccr.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 29.Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, et al. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–5. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nusse R, Fuerer C, Ching W, Harnish K, Logan C, Zeng A, et al. Wnt signaling and stem cell control. Cold Spring Harb Symp Quant Biol. 2008;73:59–66. doi: 10.1101/sqb.2008.73.035. [DOI] [PubMed] [Google Scholar]
- 31.Nusse R. Wnt signaling and stem cell control. Cell Res. 2008;18:523–7. doi: 10.1038/cr.2008.47. [DOI] [PubMed] [Google Scholar]
- 32.Fang X, Yoon JG, Li L, Yu W, Shao J, Hua D, et al. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics. 2011;12:11. doi: 10.1186/1471-2164-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagy C, Turecki G. Sensitive periods in epigenetics: bringing us closer to complex behavioral phenotypes. Epigenomics. 2012;4:445–57. doi: 10.2217/epi.12.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teodoridis JM, Strathdee G, Brown R. Epigenetic silencing mediated by CpG island methylation: potential as a therapeutic target and as a biomarker. Drug Resist Updat. 2004;7:267–78. doi: 10.1016/j.drup.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki H, Maruyama R, Yamamoto E, Kai M. DNA methylation and microRNA dysregulation in cancer. Mol Oncol. 2012 doi: 10.1016/j.molonc.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee YM, Chen HW, Maurya PK, Su CM, Tzeng CR. MicroRNA regulation via DNA methylation during the morula to blastocyst transition in mice. Mol Hum Reprod. 2012;18:184–93. doi: 10.1093/molehr/gar072. [DOI] [PubMed] [Google Scholar]
- 37.van Schooneveld E, Wouters MC, Van der Auwera I, Peeters DJ, Wildiers H, Van Dam PA, et al. Expression profiling of cancerous and normal breast tissues identifies microRNAs that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers. Breast Cancer Res. 2012;14:R34. doi: 10.1186/bcr3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Toh HC, Chow P, Chung AYF, Meyers DJ, Cole PA, et al. MicroRNA-224 is up-regulated in hepatocellular carcinoma through epigenetic mechanisms. Faseb J. 2012;26:3032–41. doi: 10.1096/fj.11-201855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veerla S, Lindgren D, Kvist A, Frigyesi A, Staaf J, Persson H, et al. MiRNA expression in urothelial carcinomas: important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int J Cancer. 2009;124:2236–42. doi: 10.1002/ijc.24183. [DOI] [PubMed] [Google Scholar]
- 40.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnold CP, Tan R, Zhou B, Yue SB, Schaffert S, Biggs JR, et al. MicroRNA programs in normal and aberrant stem and progenitor cells. Genome Res. 2011;21:798–810. doi: 10.1101/gr.111385.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim TM, Huang W, Park R, Park PJ, Johnson MD. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res. 2011;71:3387–99. doi: 10.1158/0008-5472.CAN-10-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]