Abstract
In addition to the canonical double helix, DNA can fold into various other inter- and intramolecular secondary structures. Although many such structures were long thought to be in vitro artefacts, bioinformatics demonstrates that DNA sequences capable of forming these structures are conserved throughout evolution, suggesting the existence of non-B-form DNA in vivo. In addition, genes whose products promote formation or resolution of these structures are found in diverse organisms, and a growing body of work suggests that the resolution of DNA secondary structures is critical for genome integrity. This Review focuses on emerging evidence relating to the characteristics of G-quadruplex structures and the possible influence of such structures on genomic stability and cellular processes, such as transcription.
The right-handed double helical structure of B-form DNA (B-DNA) has been known since 1953 (REF. 1). However, it has become increasingly clear that DNA can adopt a variety of alternative conformations based on particular sequence motifs and interactions with various proteins. These non-B-form secondary structures, which include G-quadruplex structures (G4 structures) (FIG. 1) as well as Z-DNA, cruciforms and triplexes (BOX 1), were originally characterized in vitro using biophysical techniques (for example, circular dichroism2). Accumulating evidence now points towards the existence of these structures under physiologically relevant conditions, and all of them are hypothesized, or even known, to have functional roles in vivo. The current wealth of genomic data — which is enabling the evolutionary comparison of motifs that can adopt non-B-form secondary structures in vitro — and the use of structure-specific antibodies, structure-binding ligands and clever experimental techniques are driving progress in this field.
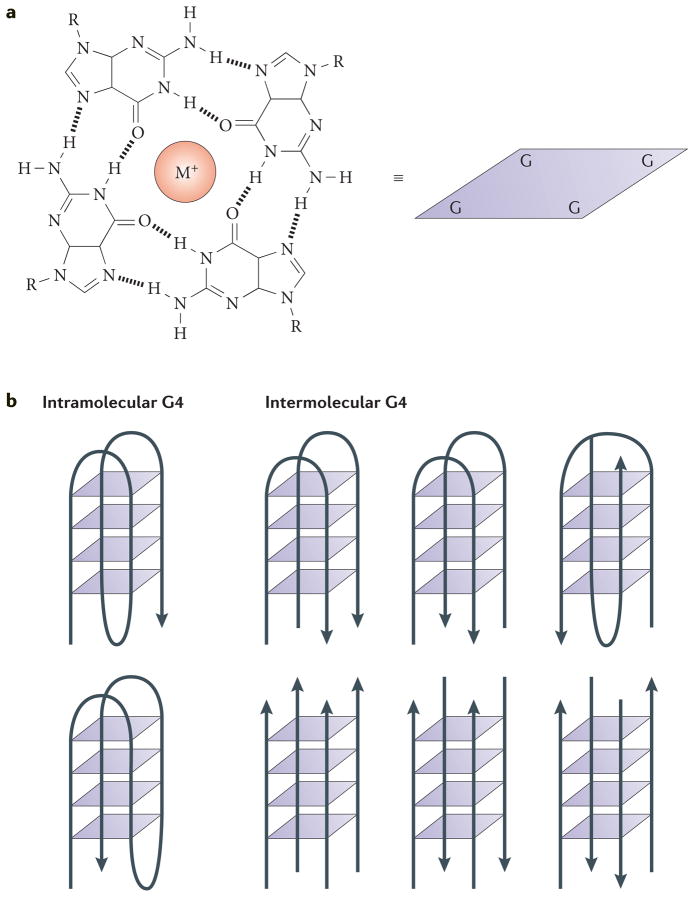
Figure 1. G-quadruplex DNA.
a | An illustration of the interactions in a G-quartet. This quartet is represented schematically as a square in the other panels of this figure. M+ denotes a monovalent cation. b | Schematic diagrams of intramolecular (left) and intermolecular (right) G-quadruplex (G4) DNA structures. The arrowheads indicate the direction of the DNA strands. The intermolecular structures shown have two (upper) or four (lower) strands.
Box 1. Other non-B-form DNA secondary structures.
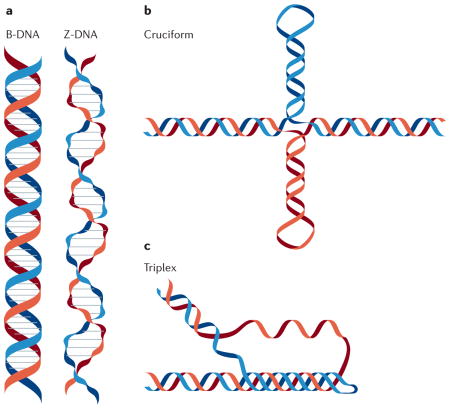
G-quadruplex (G4) structures are only one of many (ten or more) non-B-form DNA secondary structures analysed to date127. Brief descriptions of three well-studied structures are provided below.
Z-DNA
In contrast to standard B-form DNA (B-DNA), Z-DNA is a left-handed helix128 (see the figure, part a). Z-DNA motifs (that is, sequences that form Z-DNA in vitro) are tracts of alternating purines and pyrimidines, which occur about once every 3,000 bp in metazoans129. Negative supercoiling stabilizes the formation of Z-DNA under physiological salt conditions130, and it is hypothesized that Z-DNA relieves transcription-induced torsional stress131. Z-DNA motifs are tightly associated with transcriptional start sites in eukaryotic genomes132, and these motifs can also cause genome instability, although the type of damage they cause varies from prokaryotes (dinucleotide insertions and deletions) to eukaryotes (double-strand breaks resulting in larger deletions)120,121,133,134.
Cruciform structures
Negative supercoiling can also cause B-DNA to adopt a four-armed, cruciform secondary structure that resembles a Holliday junction135 (see the figure, part b). These structures require ≥6 nucleotide inverted repeats (cruciform motif) to form, and such motifs are located near replication origins, breakpoint junctions and promoters in diverse organisms136,137. In metazoans, cruciform motifs are enriched near sites of gross chromosomal rearrangements138, and deletions and translocations occur more frequently in vivo at sites of cruciform motifs than in B-DNA139–141. However, cruciforms might also serve positive roles (for example, stabilizing the human Y chromosome (reviewed in REF. 134)).
Triplex DNA
Three-stranded triplex DNA occurs when single-stranded DNA forms Hoogsteen hydrogen bonds in the major groove of purine-rich double-stranded B-DNA142 (see the figure, part c). Triplexes in which the third strand is antiparallel to the DNA duplex can form at physiological pH, and these structures are stabilized by negative supercoiling142. Sequences capable of forming triplexes are common in eukaryotes but much rarer in prokaryotes143. In mammals, triplex-forming motifs are enriched in the introns of a variety of essential genes, including those involved in development and signalling144. Additionally, triplexes are hypothesized to cause genomic instability by causing double-strand breaks that result in translocations145. However, the formation of a triplex structure in a trinucleotide repeat sequence (for example, (CAG)n) can prevent the expansion of the repeat138,139; repeat expansion is related to human genetic disorders146,147.
Although the high thermal stability of G4 structures — potentially an impediment to DNA transactions — has led to some scepticism concerning their in vivo relevance, interest in G4 structures has increased enormously in recent years owing to their unique physical properties and the presence of G-rich sequences in biologically functional regions of many genomes. For example, G-rich regions with the potential to form G4 structures (hereafter called G4 motifs) are over-represented in telomeres, mitotic and meiotic double-strand break (DSB) sites, and transcriptional start sites (TSSs; often near promoters). These findings suggest multiple roles for G4 structures. Moreover, recent work suggests that failure to resolve non-canonical DNA structures makes the sequence motifs capable of forming structural hotspots for genomic instability.
We begin this Review with an overview of G4 DNA structures, including their in vitro characterization and chromosomal locations in diverse organisms. Next, we discuss the putative roles of G4 structures at telomeres, during DNA replication, in gene regulation and in various other biological processes. Finally, we conclude by summarizing outstanding questions in the field and suggesting possible ways to address these issues.
Overview of G4 DNA
Biochemical characteristics and in vitro analyses
G4 structures are stacked nucleic acid structures that can form within specific repetitive G-rich DNA or RNA sequences (reviewed in REF. 3). In 1910, Bang4 was the first to report the fact that guanylic acid forms a gel at high concentrations, which suggested that G-rich sequences in DNA may form higher-order structures. Fifty years later, Gellert and colleagues5 used X-ray diffraction to demonstrate that guanylic acids can assemble into tetrameric structures. In these tetramers, four guanine molecules form a square planar arrangement in which each guanine is hydrogen bonded to the two adjacent guanines (that is, a G-quartet (FIG. 1a)). Stacked G-quartets form a G4 structure, and the intervening sequences are extruded as single-strand loops (although tetramolecular G4 structures may also lack loops). The sequence and size of the loop regions varies. However, loops are usually small (1–7 nucleotides (nt)), and smaller loops result in more stable G4 structures, as do longer G-tracts3. This structure is stabilized by monovalent cations that occupy the central cavities between the stacks, neutralizing the electrostatic repulsion of inwardly pointing guanine oxygens6–8.
G4 structures adopt a variety of topologies and can be classified into various groups depending on the orientation of the DNA strands (FIG. 1b). Thus, G4 structures can be parallel, antiparallel or hybrids thereof. Furthermore, they can form within one strand (intramolecular) or from multiple strands (intermolecular), and various loop structures are also possible9,10. G4 structures can be extremely stable, although the topology and stability of the G4 structure depends on many factors, including the length and sequence composition of the total G4 motif, the size of the loops between the guanines, strand stoichiometry and alignment11–13, and the nature of the binding cations14.
Chromosomal location of G4 motifs
Intramolecular G4 structures are predicted to form at specific G-rich regions in vivo that have in common a sequence motif with at least four runs of guanines (G-tracts), in which each G-tract most often contains at least three guanines (G≥3NxG≥3NxG≥3NxG≥3). G4 structures with only two stacks of guanines are possible but have low stability; here, when we refer to G4 motifs, we refer to motifs in which each G-tract contains three or more guanines. Computational analyses reveal that there are >375,000 G4 motifs in the human genome, whereas there are >1,400 G4 motifs in the Saccharomyces cerevisiae nuclear genome, including those in ribosomal and telomeric DNA, which are both particularly G4-rich15–18. Thus far, it is unclear how many of these motifs form stable G4 structures in vivo and, if they do, when they form.
Computational studies in various organisms have revealed that G4 motifs are not randomly located within genomes, but rather they tend to cluster in particular genomic regions (reviewed in REF. 19). In human, yeast and bacterial genomes, G4 motifs are similarly distributed and are over-represented in certain functional regions, such as promoters15–18,20. Furthermore, the locations and nucleotide compositions of G4 motifs are conserved in human populations and among related yeast species15,21. The nonrandom distribution of G4 motifs and the evolutionary conservation of their positions in genomes suggest that G4 motifs have one or more positive functions in the cell. In many organisms, telomeres contain a high concentration of G4 motifs owing to their high GC content and the single-stranded nature of the telomeric overhang. In diverse organisms, G4 DNA motifs are also common in G-rich micro- and minisatellites, up- and downstream of TSSs (often near promoters), within the ribosomal DNA, near transcription factor binding sites, and at preferred mitotic and meiotic DSB sites15,17,18,21,22.
G4 structures at telomeres
Telomeres are nucleoprotein complexes at the ends of linear chromosomes. They are composed of a double-stranded region and a single-stranded G-rich 3′ overhang. Telomeres are essential to protect chromosomes from degradation, end-to-end fusions, and being recognized as DSBs23. In most telomeric DNAs, guanines and cytosines are distributed asymmetrically between the two DNA strands, with the G-rich strand running 5′ to 3′ from the centromere to the telomere. For example, vertebrate telomeric DNA consists of 5′-T2AG3-3′ repeats, whereas certain ciliated protozoans such as Stylonychia lemnae have 5′-T4G4-3′ repeats. Moreover, the G-rich strand is longer than its complement, resulting in single-strand ‘G-tails’ at the very termini of chromosomes. Regardless of the precise sequence of the telomere, the G-rich strand of various telomeric sequences can usually form stable G4 structures in vitro (FIG. 2); for example, in non-denaturing polyacrylamide gels, oligonucleotides corresponding to the telomeric G-rich strand display unexpected banding patterns that are due to the formation of G4 structures6,24–26.
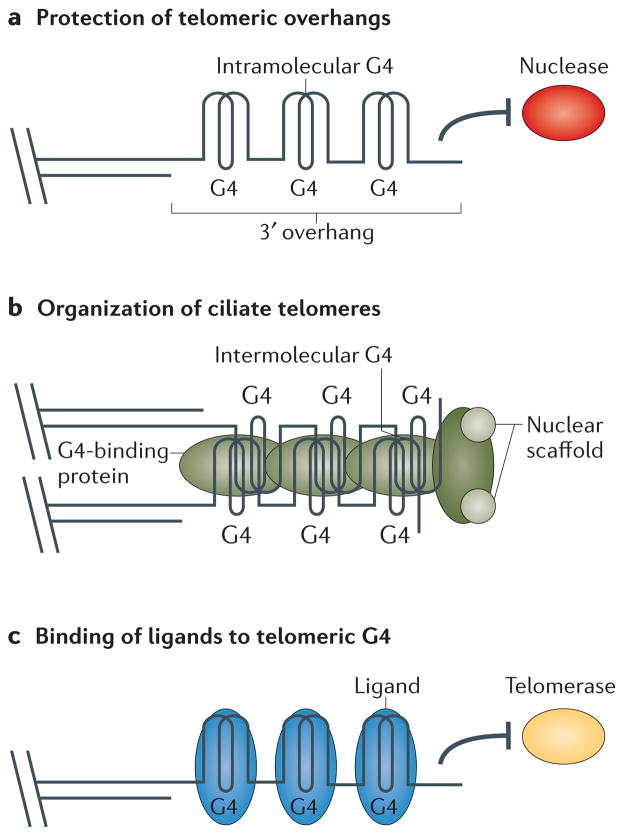
Figure 2. Putative functional roles of G-quadruplex structures at telomeres.
Telomeric sequences can fold into G-quadruplex (G4) structures in vitro. Currently, many groups are investigating the physiological relevance of this phenomenon. a | G4 structures may form at the telomeric 3′ overhang and have a role in protecting telomeres from degradation by nucleases (red) or other events. b | Work in ciliates shows that G4 structures do form at telomeres and have a role in telomere protection and tethering to the nuclear scaffold. Formation, stabilization and tethering is faciliated by G4-binding proteins (green). c | Ligands (blue) that bind to telomeric G4 structures are currently being analysed for their ability to influence telomere length by altering telomerase (yellow) activity.
Evidence for G4 structures at telomeres
The possibility that G4 structures might form in vivo is demonstrated by in vitro experiments showing that telomere structural proteins, such as TEBPα and TEBPβ in ciliates and Rap1 in S. cerevisiae, can promote the formation of G4 DNA25,27–29. By contrast, the human telomeric G-strand binding protein protection of telomeres protein 1 (POT1) promotes the unfolding of G4 structures in vitro30,31. Thus far, the most direct evidence that G4 structures exist at telomeres comes from studies in ciliates that exploit antibodies raised by ribosome display against parallel and antiparallel telomeric G4T4 structures. With these antibodies, it is possible to show that G4 structures exist in vivo at Stylonychia lemnae telomeres and to determine proteins that are required for their formation and unfolding28,32,33. Only the antibodies raised against antiparallel G4 structures bind to S. lemnae telomeres, indicating that antiparallel, and not parallel, G4 DNA is present in vivo32. In addition, several in vivo control experiments demonstrated that the anti-G4 antibodies do not induce the formation of G4 structures. The visualization of the regulation of G4 structures is an important observation because unresolved G4 structures are likely to be an obstacle for DNA replication and telomere elongation. Accordingly, telomeric G4 structures, which are present during most of the S. lemnae cell cycle, are resolved during DNA replication32. Further analysis using RNAi to silence gene expression indicates that the formation of telomeric G4 structures is dependent on two telomere binding proteins: TEBPα and TEBPβ. TEBPα binds to the telomeric overhang and recruits TEBPβ, which is able to promote the formation of G4 structures with its highly charged carboxyl terminus, as shown in vitro27,28.
As stated above, G4 structures are not present at S. lemnae telomeres during S phase. In vitro and in vivo studies demonstrate that G4 unfolding is dependent on at least three conditions. First, TEBPβ, which is essential for the formation of G4 structures, must be removed from the telomeres. This removal happens during DNA replication and requires phosphorylation of TEBPβ. Second and third, immunofluorescence and gene knockdown analyses show that two enzymes, the telomerase holoenzyme and a RecQ family helicase, are recruited to telomeric G4 structures at the end of S phase and are essential for the unfolding of telomeric G4 structures28,33–35. Currently, it is not clear how or why telomerase is needed to unwind G4 structures during DNA replication nor whether this regulation is conserved among other organisms. However, RecQ helicases in other organisms, such as Sgs1 in S. cerevisiae and WRN and BLM in humans, also act on telomeres and can unwind G4 structures in vitro (reviewed in REF. 36). To date, no-one has isolated antibodies against the human telomeric G4 structure, but the fact that TEBP homologues exist in vertebrates suggests that similar mechanisms might exist in higher eukaryotes.
There is also evidence for G4 DNA at telomeres in human cultured cells: BMVC (3,6-bis(1-methyl-4- vinylpyridinium) carbazole diiodide) is a fluorescent biomarker that binds and stabilizes G4 structures in vitro, and in vivo staining with BMVC marks the distal ends of metaphase chromosomes in human lung adenocarcinoma cells37,38, suggesting telomeric binding. However, it is not clear whether this ligand detects G4 structures formed in vivo or whether it induces G4 DNA formation. Additional in vivo experiments are required to prove the specificity of such ligands.
Possible consequences of G4 structures at telomeres
Owing to the biochemical properties of DNA polymerases, they cannot replicate the very ends of linear chromosomes. In most organisms, telomerase, a telomere-dedicated reverse transcriptase, uses its RNA subunit as a template to lengthen the G-strand of the telomere. Human telomerase is inactive in most somatic cells but is upregulated in most cancers, in which it is thought to promote the lifespan of malignant cells39. G4 structures influence telomerase activity: intramolecular antiparallel G4 structures block telomerase activity, whereas intermolecular parallel G4 DNA is permissive for extension by telomerase40–42.
Because telomerase is active in most human cancers and this activity can be influenced by G4 structures, a variety of small molecule ligands with different specificities and target regions that bind and stabilize G4 structures are being tested in various assays43. The hope is that ligands that promote the formation of certain types of telomeric G4 structures might inhibit telomerase by preventing annealing of telomerase RNA to G-strand overhangs. For example, telomestatin has nanomolar affinity for telomeric G4 structures (which is nearly two orders of magnitude lower than its affinity for double-stranded DNA) and stabilizes intramolecular antiparallel G4 structures in vitro44,45. Moreover, telomestatin inhibits telomerase46 and causes gradual telomere shortening and growth arrest or apoptosis in human tissue culture cancer cells47–52. However, telomeric DNA damage also increases in telomestatin-treated cells50,53,54. Thus, telomere shortening in telomestatin-treated cells might also be due to capping defects, especially as telomere repeat binding factor 2 (TRF2) and POT1 telomere binding are lost in these cells. Indeed, in S. cerevisiae, G4 structures are thought to contribute to telomere capping when natural capping is impaired55. Further research is required to determine whether G4 ligands are effective in vivo, whether they are specific for telomeric DNA and whether their presence has deleterious effects on non-telomeric G4 structures.
Effects of G4 structures on DNA replication
During DNA replication, the two strands of the DNA double helix are separated by the replicative helicase: one strand serves as the template for leading strand synthesis and the other for lagging strand synthesis. Although leading strand DNA replication can be continuous, the lagging strand is replicated discontinuously, making it transiently single-stranded; this is a conformation that provides opportunities for G4 structure formation. Thus, during DNA replication, G4 structures may form inappropriately, especially on the lagging strand template (FIG. 3), and this formation is more likely to occur when DNA replication is slowed. In addition, some G4 structures could be present during DNA replication because they have roles in transcriptional regulation (see below). Whether G4 structures are pre-existing or form during DNA replication, they must be resolved for completion of DNA replication because the sequence comprising the G4 structure cannot serve as a template until it is unfolded. Thus, helicases are likely to be necessary to unwind G4 structures.
Figure 3. Putative functional roles of G-quadruplex structures during DNA replication.
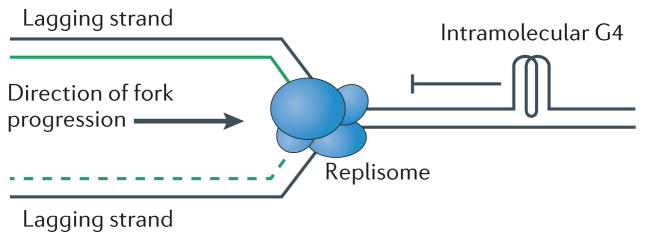
Computational studies show that in all tested organisms, many regions in the genome have the ability to form G-quadruplex (G4) structures. In vitro and in vivo studies indicate that unresolved G4 structures may influence DNA replication by slowing or stalling the replication fork machinery (replisome; blue).
We surveyed the literature and found that 22 different helicases have been tested for their ability to bind and/or unwind G4 structures in vitro, and all but one, the Escherichia coli RecBCD helicase, was positive (K.P., M.L.B. and V.A.Z., unpublished observations; summarized in Supplementary information S1 (table)). These data suggest that G4 unwinding is a non-specific activity of many DNA helicases. However, most of these unwinding studies are qualitative, and it is difficult to ascertain from them whether a given helicase is particularly effective at unwinding G4 structures and/or whether G4 structures are a preferred substrate for that helicase. Most of the human helicases that unwind G4 structures in vitro56–60 are associated with human diseases that cause genomic instability, including the RecQ helicases WRN (associated with premature ageing) and BLM (associated with increased cancer risk) as well as FANCJ (associated with increased cancer risk) and PIF1 (associated with increased cancer risk). The best evidence that human disease is associated with loss of G4 unwinding comes from the finding that cell lines from human patients with Fanconi anaemia carrying FANCJ mutations display deletions that overlap G-rich regions with the potential to form G4 structures56. In addition, telomestatin, a chemical ligand that is able to stabilize G4 structures in vitro53,61,62, causes impaired proliferation and increased apoptosis and DNA damage in FANCJ-deficient cells63. The association of these helicases with inherited genome instability has heightened interest in the possibility that G4 unwinding might suppress both premature ageing and cancer by regulating G4 structures.
Some enzymes are far more active on G4 structures than others. The S. cerevisiae Pif1 helicase acts at G4 motifs64, and members of the Pif1 DNA helicase family are particularly efficient in vitro unwinders of parallel intramolecular G4 substrates59. Pif1 is a multi-functional DNA helicase that binds >1,000 sites in the genome of mitotic cells, of which ~10% overlap G4 motifs, which represents ~25% of the G4 motifs in this organism. Twenty-five per cent is likely to be an underestimate as, for technical reasons, this number excludes the large number of G4 motifs in ribosomal and telomeric DNA, both of which are strong Pif1 binding sites64. Several genetic assays show that in the absence of Pif1, DNA replication slows and DSBs occur at many of the G4 motifs that are normally bound by Pif1. G4 motifs also show a high mutation rate in Pif1-deficient cells, and these mutations eliminate the ability of the motif to form a G4 structure without necessarily reducing the high GC content of the motif. When these mutated motifs are put back in the genome, they no longer bind Pif1, slow DNA replication or cause DSBs. Together, these data make a strong argument that G4 structures form in vivo and that their resolution by Pif1 suppresses genome instability64. Other studies also found instability of G4 motifs in pif1 cells59,65. This instability was particularly pronounced when the G4 motifs were on the template for leading strand synthesis, but this result may reflect the repetitive nature of the G4 substrate used in this analysis. The frequent mutation of G4 motifs in pif1 mutant cells suggests the involvement of error-prone processes when G4 motifs are replicated and repaired in Pif1-deficient cells64. Indeed, in DT40 chicken cells, REV1, a translesion polymerase, is implicated in replication fork progression past G4 motifs on the leading strand66.
There are also suggestions that human PIF1 acts at G4 motifs. One study used chromatin immunoprecipitation followed by sequencing (ChIP–seq) in combination with in vivo labelling with pyridostatin, a G4 binding molecule67. Genome-wide, pyridostatin bound preferentially to G4 motifs, where it caused replication and transcription-dependent damage that was detected by its high γH2Ax content. Many of the γH2Ax foci overlap with GFP–PIF1 foci in the pyridostatin-treated human cells. The current hypothesis is that G4 formation or stabilization blocks transcription and/or replication, resulting in DNA damage.
Similar to what is seen in cells from patients with Fanconi anaemia whose disease is due to mutations in the FANCJ helicase, mutations in the Caenorhabditis elegans DOG-1 helicase, which is distantly related to FANCJ, cause genome-wide deletions in G-rich sequences with the potential to form G4 structures68,69. The mutation rate in dog-1 mutants is very high (up to 4% per generation68) and increases with the length of the G-tract69. Finally, the activity of regulator of telomere elongation helicase (RTEL) family helicases is also hypothesized to be directed towards G4 structures. Recent data indicate that the human RTEL helicase helps to resolve G4 DNA at telomeres, perhaps in conjunction with BLM, to ensure telomere stability70. Although biochemical evidence of G4 unwinding is lacking for RTEL homologues from other organisms, current data indicate that they may function similarly to human RTEL. For instance, C. elegans rtel-1 has high sequence similarity to dog-1, although G-rich sequences are not unstable in worms deficient for rtel-1 (REF. 71) as they are in dog-1 mutant animals. However, mutation of rtel-1 and him-6 (a BLM homologue) is synthetically lethal in C. elegans, suggesting that RTEL-1 may function in concert with one or more additional helicases (DOG-1 and/or HIM-6) to resolve G4 structures.
G4 structures in transcription
The high concentration of G4 motifs near promoter regions suggests a potential function of G4 structures in gene regulation. Indeed, one or more G4 motifs are found within 1,000 nt upstream of the TSS of 50% of human genes72. Intriguingly, bioinformatics show that the promoters of human oncogenes and regulatory genes (for example, transcription factors) are more likely than the average gene to contain G4 motifs, whereas G4 motifs are under-represented in the promoters of housekeeping and tumour suppressor genes22,72. A similar enrichment of G4 motifs in promoter regions is found in other organisms, including yeast, plants and bacteria15,17,20,73,74. Additionally, in humans, G4 motifs are less often found in the template strand than in the non-template strand. Those that are on the template strand tend to cluster at the 5′ end of the 5′UTR75. In yeast, there is no distinct asymmetry in G4 motif location between the non-template and template strands, but there is a correlation between nucleosome-free regions and G4 motifs in promoters15, a finding that supports the prediction that G4 structures will form more easily in nucleosome-free regions17. Experiments in bacteria using a G4 motif on the non-template strand of a plasmid-borne transcribed gene demonstrate loop formation on the opposite strand of the G4 motif, suggesting the existence of G4 structures that form upon transcription in living cells76. Such structures may help to keep the transcribed template accessible for transcription by preventing it annealing to its complementary strand. In this way, G4 structures could contribute to high transcription levels of certain genes (FIG. 4).
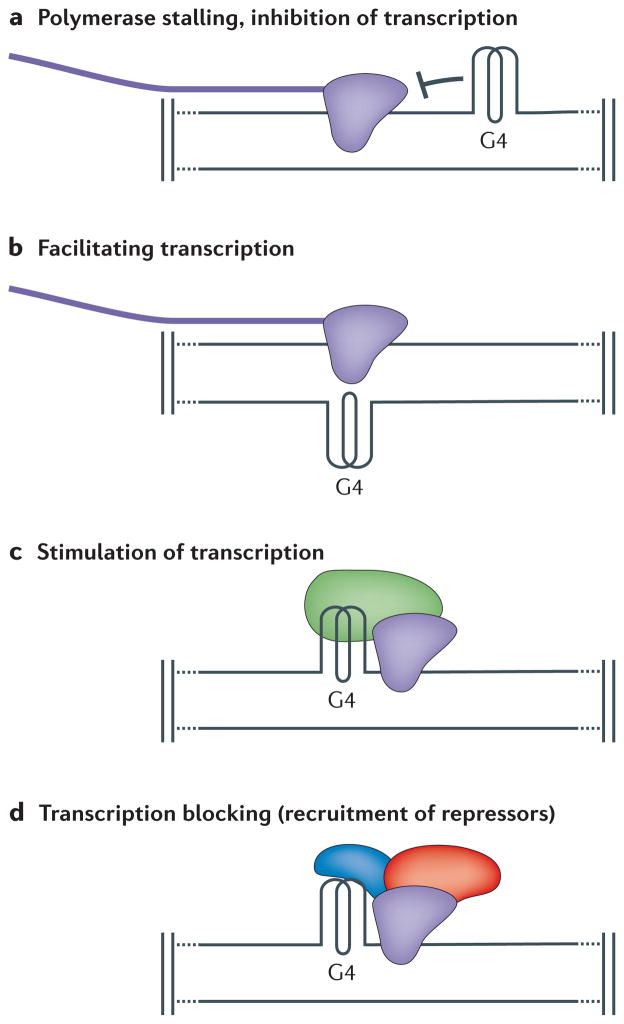
Figure 4. Putative functional roles of G-quadruplex structures during transcription.
Genome-wide bioinformatic analyses identified loci with high potential to form G-quadruplex (G4) structures. Among these loci, the promoters, transcription factor binding sites and 5′UTR regions of mRNAs are highly enriched for G4 motifs. These analyses, together with protein–G4 interaction studies, provide insights into predicted functions of G4 structures during transcription. a | G4 structures are postulated to block transcription by inhibiting polymerase (purple). b | G4 structures are postulated to facilitate transcription by keeping the transcribed strand in the single-stranded conformation. c | G4 DNA may stimulate transcription by recruiting proteins (green) that recruit or stimulate polymerase. d | G4 structures are suggested to block transcription via the recruitment of G4 binding proteins (blue), which directly or indirectly (red) repress transcription.
Possible consequences of G4 structures formed during transcription
It is well known that supercoiling has both positive and negative effects on transcription77, and G4 structures are thought to form as a result of supercoiling- induced stress during transcription78. In vitro studies show that the formation of G4 structures can compensate for the negative supercoiling78,79. These findings suggest that G4 structures in or near promoter regions may influence transcription in both positive and negative ways (FIG. 4). First, depending on which DNA strand encodes the G4 motif, the structure could either inhibit transcription (if the motif is on the template strand, blocking the transcription machinery) or enhance transcription (if the motif is on the non-template strand, maintaining the transcribed strand in a single-stranded conformation). Second, proteins bound to the G4 structures (for example, transcriptional enhancers versus repressors) could also affect transcription (reviewed in REF. 80).
One of the best-studied systems for a role of G4 structures in transcription involves the mammalian MYC (also known as c-MYC) locus (reviewed in REFS 3,79), although findings similar to those discussed below have been reported for multiple loci80–84. MYC is a transcription factor whose expression is associated with cell proliferation. Increased levels of MYC expression are observed in 80% of human cancer cells, and this increase promotes tumorigenesis85–90. Nuclease hypersensitive element III1 (NHE III1), which is downstream of the MYC promoter, controls >80% of the MYC transcription. This element contains a G4 motif that forms a G4 structure in vitro91. Footprinting studies and luciferase reporter assays comparing the expression of a gene with a wild-type NHE III1 versus one with a mutated NHE III1 that cannot form a G4 structure demonstrate that the G4 motif in NHE III1 represses transcription92. In another study, TMPyP4, a compound that binds to and stabilizes G4 structures (but also binds duplex DNA)93,94, reduced MYC transcription in lymphoma cell lines and showed antitumour activity in mice92,95. This reduction is speculated to be mediated by TMPyP4 binding to the G4 structure in NHE III1 of MYC. However, given that TMPyP4 binding is not limited to G4 structures and the many G4 motifs in the genome, more analysis is required to determine its mechanism of action. GQC-05, an analogue of ellipticine (an antineoplastic drug), is another promising therapeutic ligand. GQC-05 binds the G4 structure in the NHE III1 region of MYC in vitro with high affinity and selectivity, and when added to Burkitt’s lymphoma cell lines, GQC-05 results in reduced levels of transcribed MYC mRNA96. However, a recent publication found that 11 known G4 DNA ligands that affect MYC expression in cell-free assays do not interact directly with the MYC G4 structure in certain Burkitt’s lymphoma cell lines97, clouding the interpretation of the GQC-05 results.
Nucleolin, the most abundant nucleolar phosphoprotein in eukaryotic cells, is also proposed to regulate MYC transcription via its interaction with NHE III1. This hypothesis is based on the in vivo binding of nucleolin to the MYC promoter in HeLa cells and the dose-dependent reduction in MYC transcription that occurs in nucleolin-treated cells98. One hypothesis is that nucleolin-mediated G4 formation in NHE III1 inhibits MYC transcription by masking binding sites for MYC transcriptional activators, such as the transcripton factor SP1 and cellular nucleic acid-binding protein (CNBP)99. However, human nucleolin binds many G4 structures and can induce the formation of G4 DNA in vitro98,100–103. Thus, more work is needed to establish that nucleolin-associated changes in MYC transcription are a direct result of its effects on G4 structure formation within the NHE III1 element.
Regulation through proteins binding to G4 structures
Transcription may also be altered by G4 binding proteins that affect the formation and unfolding of G4 structures. For example, myosin D (MyoD) family proteins are transcription factors that bind to E-boxes in the promoters of several muscle-specific genes to regulate muscle development104. In vitro, MyoD homodimers bind preferentially to G4 structures that are derived from the promoter sequences of muscle specific genes105. One hypothesis is that when G4 structures form in the promoters of E-box driven genes, MyoD homodimers preferentially bind to the G4 structure and not the E-box. Consequently, MyoD–MyoE heterodimers, which cannot bind G4 structures, bind to the E-box instead and enhance gene transcription106. However, like the MYC experiments, additional work is needed to prove this hypothesis.
In addition to gene-specific approaches, results from genome-wide studies analysing the effects of drugs that stabilize and/or induce G4 formation have been used to argue that G4 structures affect transcription79,107. Indeed, expression levels of many genes are influenced by treating cells with G4 ligands. Similar studies have investigated the effects of mutations in helicases known to unwind G4 DNA on transcription genome wide17,108. For instance, in human fibroblasts deficient for the WRN or BLM RecQ helicases, the transcription of genes that are predicted to form intramolecular G4 structures is significantly upregulated (P < 0.0001), and this upregulation correlates with the G4 motifs, not simple G-richness108. The genes associated with G4 motifs account for 20–30% of all transcripts that are upregulated in WRN and BML mutant cells.
Although such studies support a role for G4 structures in transcription, when interpreting genome-wide studies the possibility must be considered that many of the observed changes in gene expression may be indirect. However, in diverse organisms, genes whose expression is affected by G4 ligands are statistically associated with the presence of nearby G4 motifs, which provides some of the best evidence for widespread effects of G4 structures on transcription.
A general criticism of models in which G4 structures affect transcription is that G4 formation is too slow and the stability of G4 structures is too high for them to be used as regulatory elements. This criticism can also be raised against hypotheses suggesting that G4 structures affect telomeres or DNA replication. Indeed, it is well documented that intermolecular G4 DNA structures form and resolve slowly under physiological conditions109,110. However, the existence of chaperones (for example, TEBPβ and Rap1) that promote the formation of G4 DNA27–29 suggests that nature has evolved mechanisms to overcome this slow formation. A recent thermodynamic and kinetic measurement of G4 structure formation indicates that G4 structures can form cooperatively111. Rates of formation for intramolecular G4 structures have also been reported for human telomeric G4 DNA (millisecond timescale)112, and it is possible that other intramolecular G4 structures form as readily. This possibility is simple to test and should be demonstrated directly for other G4 motifs that are proposed to form intramolecular G4 structures that function in vivo. Unwinding of G4 structures in a timely manner can also no longer be considered a problem given the discovery of helicases that bind and unwind G4 motifs with high efficiency (see above).
Other roles for G4 DNA
Epigenetic regulation
A new hypothesis suggests that G4 structures might influence epigenetic regulation of gene expression. Maintaining epigenetic marks, such as histone methylation, is essential for stable gene expression and cell identity, and these marks must therefore be preserved after DNA replication and repair. As reported above, G4 structures are thought to cause replication fork stalling. These stalled forks might be restarted with the aid of translesion polymerases, as suggested by data from DT40 chicken cells66, in which REV1, a Y family translesion polymerase113, is implicated in G4 lesion bypass. In the absence of REV1, DNA synthesis is uncoupled from histone recycling mechanisms, and transcriptional activation is blocked66. The authors postulate that REV1 functions in replication at G4 motifs in order to preserve histone modifications66. A recent publication extends this work by showing by microarray analysis that lack of REV1 causes genome-wide dys-regulation of G4-dependent transcription in DT40 cells (P value = 0.005), and this dysregulation is worsened by mutation of the WRN, BLM and FANCJ helicases114.
Origins of replication
It is well documented that chromatin can influence the timing of origin activation during DNA replication115. Recently, genome-wide analysis of replication origins116 using a short nascent strand sequencing approach together with deep sequencing techniques identified a large number of new origins in different human cell types. Most of the identified peaks overlap with previously identified origins; however, many of the newly identified origins are significantly associated with G4 motifs. The authors propose that G4 structures near origins promote origin of replication complex binding and thereby influence origin activation116, although direct proof for this model is not yet available.
Meiosis
G4 structures are also suggested to be involved in the alignment of sister chromatids during meiosis. One hypothesis is that G4 structures assist in the formation of the telomere-dependent bouquet structure during meiosis (FIG. 5a), but there is no direct evidence for this appealing possibility26 Various G4-promoting proteins (FIG. 5a, pink) might be involved in formation of the G4 and tethering of the bouquet. G4 structures are also proposed to have a more general role in meiosis: for example, by promoting meiotic homologous recombination76,117 (FIG. 5b). This idea is supported by genome-wide computational studies in yeast that demonstrate overlap between G4 motifs and preferred meiotic DSB sites15, but Spo11, the enzyme that makes the DSBs, does not cleave at G4 motifs118. However, a role for G4 DNA in meiosis is supported by the finding that the S. cerevisiae Hop1 protein, which is a major component of the chromosome axial element–synaptonemal complex during meiosis, promotes G4 formation in vitro119,120. The multifunctional protein Kem1 also binds G4 structures in vitro and cleaves in the single stranded region 5′ of the G4 structures. Together with the fact that kem1Δ cells arrest during meiotic prophase, these results led to speculation that Kem1 acts on G4 structures in vivo121. In addition, the MRX complex, which is composed of Mre11, Rad50 and Xrs2 and acts during meiotic DSB formation, has a high affinity for G4 structures in vitro122,123. However, there is not yet in vivo evidence that Hop1, Kem1 or the MRX complex carry out their meiotic functions by acting at G4 structures.
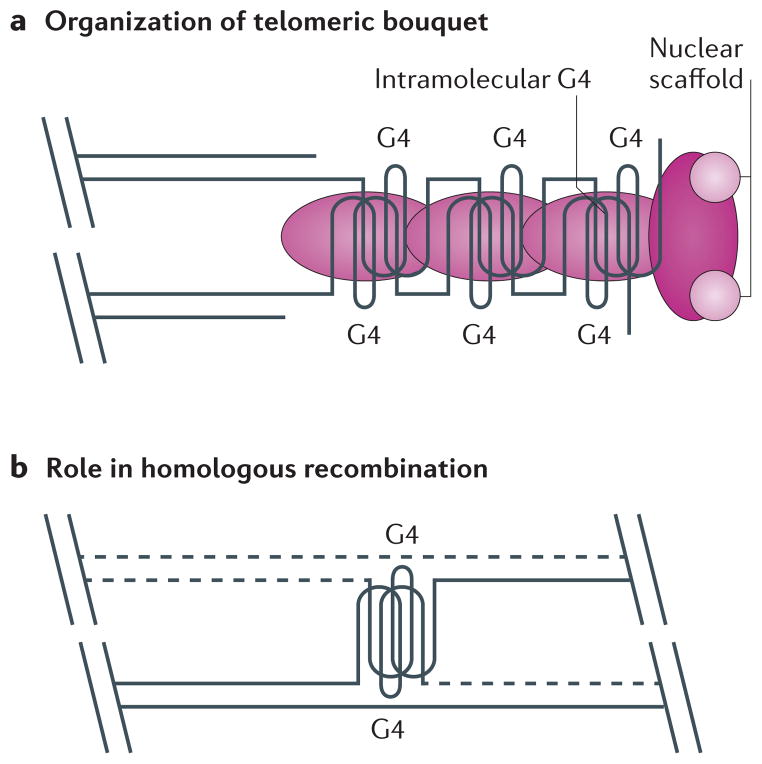
Figure 5. Putative roles for G-quadruplex structures in meiosis.
a | It has been proposed that G-quadruplex (G4) structures might assist in the formation of the telomere-dependent bouquet structure during meiosis26. G4-promoting proteins (pink) could be involved in the formation of G4 structures and tethering of the telomeric bouquet to the nuclear scaffold. b | It has also been suggested that G4 structures could promote meiotic homologous recombination76,117 if there is overlap between G4 motifs and preferred meiotic double-strand break sites15. Sites of homologous recombination are indicated by the dashed lines.
Recombination
In several pathogenic microorganisms, recombination provides the basis for antigenic variation in which the pathogen escapes its host’s immune surveillance by changing the identity of a surface antigen. There is good evidence that Neisseria gonorrhoeae, the bacterium that causes human gonorrhoea, uses a G4 based system to regulate expression of the genes that allow it to avoid the human immune system124. N. gonorrhoeae encodes many pilin genes, the products of which make up the hair-like projections, called pili, on the bacterium’s surface. However, only the gene in the pilE locus is expressed, and the identity of the gene at this site switches among the different pilin genes by a recombinational mechanism. The region upstream of the pilE locus contains a 12 bp G-rich segment that is required for antigenic variation and that can form a parallel intramolecular G4 structure in vitro. Mutations that eliminate antigenic variation in vivo also eliminate the ability of the segment to form a G4 structure, while mutations in the loop region of the structure affect neither antigenic variation nor G4 structure formation. Moreover, treating cells with the G4 ligand N-methyl mesoporphyrin IX affects pilE gene conversion events. The N. gonorrhoeae RecQ helicase is one of several proteins required in trans for efficient antigenic variation, providing additional evidence for a role for RecQ helicases at G4 structures in vivo. G4-based N. gonorrhoeae pilE recombination is perhaps the best evidence for a functional role of G4 DNA.
Conclusions
Although different in their three-dimensional conformation, G4 structures and the other non-B-form DNA secondary structures included in BOX 1 display some similarities. First, they can all form readily under the proper in vitro conditions. Second, formation of all of these secondary structures can help to relax negative DNA supercoiling, and Hoogsteen base pairing is often involved in stabilizing the structures. Third, the evolutionary conservation of the motifs capable of forming these secondary structures and the cellular machinery available to resolve them (for example, helicases and mismatch repair) argues for their existence in vivo. However, although they are of considerable interest from a chemical standpoint, some chromosome biologists remain sceptical that these secondary structures are physiologically relevant. G4 DNA provides an excellent example of the gulf between the wealth of in vitro data and the relative scarcity of results demonstrating formation and function of these structures in vivo. The findings that G4 motifs are evolutionarily conserved, over-represented in certain regions and associated with a specific subset of genomic features provides good, albeit indirect, evidence for G4 structures in vivo.
Direct evidence for G4 structures in vivo has been slow in coming. G4-specific antibodies and ligands provide support for G4 DNA in vivo, especially at telomeres, but it is difficult to demonstrate convincingly that the specificity of these reagents is high enough to rule out the possibility that their effects are due to association with B-DNA. Genetic experiments provide the most persuasive evidence to date for the in vivo existence of G4 structures during replication64,68,69 and transcription99. Regardless of the process or function in question, one must test directly for positive roles of G4 structures, for instance by mutating G4 motifs in promoter regions or meiotic DSB sites and determining whether loss of the ability to form a G4 structure affects downstream processes. However, in the end, the most convincing evidence for the existence of G4 structures in vivo will be a direct demonstration of these structures in vivo. Doing so will require a creative approach to isolate the structures with sufficient purity that they can be characterized by the kinds of approaches used to analyse in vitro-generated G4 structures.
To summarize, G4 motifs are ubiquitous in prokaryotic and eukaryotic genomes, and their location is often conserved in closely related species. These motifs may form G4 structures in vivo, and the G4 structures may have functional roles, such as regulating recombination, meiotic DSB formation and/or transcription or providing a template for an RNA that forms a G4 structure that affects its post-transcriptional behaviour (see below). Alternatively (or in addition), G4 DNA formation may be pathological, occurring only occasionally owing to a problem in DNA mechanics, such as slowed DNA replication (as in the presence of hydroxyurea), which would provide more time for G4 DNA formation, especially during lagging strand replication. Pathological G4 structures could form at sites where G4 DNA has a direct or indirect function (for example, meiotic DSB sites in mitotic cells) or at sites that are complementary to an RNA containing a G4 structure that has a function in the RNA (in this case, the G4 RNA has a function but its complement in the DNA does not). Although this Review concerns DNA secondary structures, we would be remiss without noting that similar structures, especially G4 structures, can form in RNA. One possibility is that G4 motifs are encoded in the DNA but mainly function at the RNA level. G4 RNA structures are reported to affect mRNA splicing, translation and degradation (reviewed in REFS 8,125,126). It seems clear that the study of non-canonical RNA and DNA secondary structures will provide fertile ground for research for the foreseeable future.
Supplementary Material
Acknowledgments
We thank the US National Institutes of Health, the American Cancer Society and the German Research Organization (DFG) for support.
Glossary
- B-form DNA
(B-DNA). The canonical right-handed double helical secondary structure assumed by bulk DNA in vivo
- Non-B-form secondary structures
Any DNA secondary structure that differs from B-form DNA. Such structures are likely to arise at defined sequence motifs owing to local factors acting on the B-form DNA
- G-quadruplex structures
(G4 structures). Stable DNA secondary structures that can form from motifs containing tracts of tandem guanines. The guanines hydrogen bond in a planar arrangement, forming stacks connected by single-stranded DNA loops. The DNA strands can be parallel or antiparallel, and the G4 structures can form intra- or intermolecularly
- Z-DNA
Left-handed helical DNA that can form from tracts of alternating purines and pyrimidines
- Cruciforms
Four-armed DNA secondary structures, similar to Holliday junctions, that can form at inverted repeat sequences and are stabilized by DNA supercoiling
- Triplexes
Three-stranded DNA in which single-stranded DNA hydrogen bonds into the major groove of purine-rich standard B-form DNA
- Telomeres
The ends of linear chromosomes, usually consisting of GC-rich repeated DNA, with guanines clustered in the strand that forms the 3′ end of the chromosome. The G-rich strand is longer than the C-rich strand so that telomeres contain both double- and single-stranded DNA. Sequence-specific binding proteins protect both duplex and single-stranded telomeric DNA from degradation, fusions and checkpoints
- Helicase
A class of enzymes that function as molecular motors, using the energy of ATP hydrolysis to unwind base-paired DNA or RNA. Helicases can also translocate along and displace proteins from nucleic acids
- γH2Ax
A phosphorylated histone H2A variant that accumulates at regions of DNA damage
- Telomere-dependent bouquet structure
A structure formed by telomeres in early meiosis. It is associated with the nuclear scaffold
- Hoogsteen base pairing
Base pairing that differs from the normal Watson–Crick base pairing
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Kypr J, Kejnovska I, Renciuk D, Vorlickova M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huppert JL. Structure, location and interactions of G-quadruplexes. FEBS J. 2010;277:3452–3458. doi: 10.1111/j.1742-4658.2010.07758.x. [DOI] [PubMed] [Google Scholar]
- 4.Bang I. Untersuchungen über die Guanylsäure. Biochem Z. 1910;26:293–231. (in German) [Google Scholar]
- 5.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. This is the first observation that guanylic acids can assemble into higher-order structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. This study demonstrates that oligonucleotides composed of cilate telomeric repeat sequences form G-quartets in the presence of certain monovalent cations (for example, Na+ and K+). The authors also propose that G-quartets may form at telomeres in vivo and must be dealt with by the replication machinery. [DOI] [PubMed] [Google Scholar]
- 7.Wilson WD, Sugiyama H. First international meeting on quadruplex DNA. ACS Chem Biol. 2007;2:589–594. doi: 10.1021/cb7001686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong HM, Payet L, Huppert JL. Function and targeting of G-quadruplexes. Curr Opin Mol Ther. 2009;11:146–155. [PubMed] [Google Scholar]
- 9.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazel P, Parkinson GN, Neidle S. Predictive modelling of topology and loop variations in dimeric DNA quadruplex structures. Nucleic Acids Res. 2006;34:2117–2127. doi: 10.1093/nar/gkl182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardin CC, Perry AG, White K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers. 2000;56:147–194. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 12.Guedin A, Gros J, Alberti P, Mergny JL. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bugaut A, Balasubramanian S. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry. 2008;47:689–697. doi: 10.1021/bi701873c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capra JA, Paeschke K, Singh M, Zakian VA. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput Biol. 2010;6:e1000861. doi: 10.1371/journal.pcbi.1000861. This study reports a genome-wide computational analysis identifying the location and evolutionary conservation of G4 motifs in S. cerevisiae and related yeasts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hershman SG, et al. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. The authors performed a genome-wide computational analysis that identified all regions in the human genome with a high potential to form G4 structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huppert JL. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem Soc Rev. 2008;37:1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- 20.Rawal P, et al. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakken S, Rognes T, Hovig E. The disruptive positions in human G-quadruplex motifs are less polymorphic and more conserved than their neutral counterparts. Nucleic Acids Res. 2009;37:5749–5756. doi: 10.1093/nar/gkp590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zakian VA. Telomeres: the beginnings and ends of eukaryotic chromosomes. Exp Cell Res. 2012;318:1456–1460. doi: 10.1016/j.yexcr.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson E, et al. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987;51:899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- 25.Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. This in vitro analysis demonstrates that telomeric DNA can fold into G4 structures. [DOI] [PubMed] [Google Scholar]
- 26.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 27.Fang G, Cech TR. The β subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell. 1993;74:875–885. doi: 10.1016/0092-8674(93)90467-5. [DOI] [PubMed] [Google Scholar]
- 28.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nature Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. This study includes compelling evidence for the in vivo existence of G4 structures at telomeres. Telomere-binding proteins are shown to regulate the formation of such structures. [DOI] [PubMed] [Google Scholar]
- 29.Giraldo R, Rhodes D. The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. EMBO J. 1994;13:2411–2420. doi: 10.1002/j.1460-2075.1994.tb06526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci USA. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Nora GJ, Ghodke H, Opresko PL. Single molecule studies of physiologically relevant telomeric tails reveal POT1 mechanism for promoting G-quadruplex unfolding. J Biol Chem. 2011;286:7479–7489. doi: 10.1074/jbc.M110.205641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaffitzel C, et al. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paeschke K, et al. Telomerase recruitment by the telomere end binding protein-β facilitates G-quadruplex DNA unfolding in ciliates. Nature Struct Mol Biol. 2008;15:598–604. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- 34.Postberg J, Tsytlonok M, Sparvoli D, Rhodes D, Lipps HJ. A telomerase-associated RecQ protein-like helicase resolves telomeric G-quadruplex structures during replication. Gene. 2012;497:147–154. doi: 10.1016/j.gene.2012.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Juranek SA, Paeschke K. Cell cycle regulation of G-quadruplex DNA structures at telomeres. Curr Pharm Des. 2012;18:1867–1872. doi: 10.2174/138161212799958404. [DOI] [PubMed] [Google Scholar]
- 36.Paeschke K, McDonald KR, Zakian VA. Telomeres: structures in need of unwinding. FEBS Lett. 2010;584:3769–3772. doi: 10.1016/j.febslet.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Q, et al. Verification of specific G-quadruplex structure by using a novel cyanine dye supramolecular assembly: I. recognizing mixed G-quadruplex in human telomeres. Chem Commun. 2009;9:1103–1105. doi: 10.1039/b820101c. [DOI] [PubMed] [Google Scholar]
- 38.Chang CC, et al. A novel carbazole derivative, BMVC: a potential antitumor agent and fluorescence marker of cancer cells. Chem Biodivers. 2004;1:1377–1384. doi: 10.1002/cbdv.200490100. [DOI] [PubMed] [Google Scholar]
- 39.Shay JW, Wright WE. Role of telomeres and telomerase in cancer. Seminars Cancer Biol. 2011;21:349–353. doi: 10.1016/j.semcancer.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. The authors report the first observation that telomerase action is influenced by G4 structures. [DOI] [PubMed] [Google Scholar]
- 41.Oganesian L, Moon IK, Bryan TM, Jarstfer MB. Extension of G-quadruplex DNA by ciliate telomerase. EMBO J. 2006;25:1148–1159. doi: 10.1038/sj.emboj.7601006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oganesian L, Graham ME, Robinson PJ, Bryan TM. Telomerase recognizes G-quadruplex and linear DNA as distinct substrates. Biochemistry. 2007;46:11279–11290. doi: 10.1021/bi700993q. [DOI] [PubMed] [Google Scholar]
- 43.Neidle S. Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010;277:1118–1125. doi: 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- 44.Rezler EM, et al. Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J Am Chem Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- 45.Kim MY, Vankayalapati H, Shin-Ya K, Wierzba K, Hurley LH. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. J Am Chem Soc. 2002;124:2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 46.De Cian A, et al. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc Natl Acad Sci USA. 2007;104:17347–17352. doi: 10.1073/pnas.0707365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gomez D, et al. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004;32:371–379. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim MY, Gleason-Guzman M, Izbicka E, Nishioka D, Hurley LH. The different biological effects of telomestatin and TMPyP4 can be attributed to their selectivity for interaction with intramolecular or intermolecular G-quadruplex structures. Cancer Res. 2003;63:3247–3256. [PubMed] [Google Scholar]
- 49.Shammas MA, et al. Telomerase inhibition and cell growth arrest after telomestatin treatment in multiple myeloma. Clin Cancer Res. 2004;10:770–776. doi: 10.1158/1078-0432.ccr-0793-03. [DOI] [PubMed] [Google Scholar]
- 50.Tahara H, et al. G-quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3′ telomeric overhang in cancer cells. Oncogene. 2006;25:1955–1966. doi: 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- 51.Tauchi T, et al. Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: involvement of ATM-dependent DNA damage response pathways. Oncogene. 2003;22:5338–5347. doi: 10.1038/sj.onc.1206833. [DOI] [PubMed] [Google Scholar]
- 52.Tauchi T, et al. Telomerase inhibition with a novel G-quadruplex-interactive agent, telomestatin: in vitro and in vivo studies in acute leukemia. Oncogene. 2006;25:5719–5725. doi: 10.1038/sj.onc.1209577. [DOI] [PubMed] [Google Scholar]
- 53.Gomez D, et al. Interaction of telomestatin with the telomeric single-strand overhang. J Biol Chem. 2004;279:41487–41494. doi: 10.1074/jbc.M406123200. [DOI] [PubMed] [Google Scholar]
- 54.Gomez D, et al. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006;66:6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- 55.Smith JS, et al. Rudimentary G-quadruplex-based telomere capping in Saccharomyces cerevisiae. Nature Struct Mol Biol. 2011;18:478–485. doi: 10.1038/nsmb.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.London TB, et al. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J Biol Chem. 2008;283:36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mohaghegh P, et al. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ribeyre C, et al. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanders CM. Human Pif1 helicase is a G-quadruplex DNA binding protein with G-quadruplex DNA unwinding activity. Biochem J. 2010;430:119–128. doi: 10.1042/BJ20100612. [DOI] [PubMed] [Google Scholar]
- 61.Arola A, Vilar R. Stabilisation of G-quadruplex DNA by small molecules. Curr Top Med Chem. 2008;8:1405–1415. doi: 10.2174/156802608786141106. [DOI] [PubMed] [Google Scholar]
- 62.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr Opin Struct Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 63.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. This paper demonstrates that the S. cerevisiae Pif1 helicase binds to G4 motifs genome-wide and is important for DNA replication and genome stability at such sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lopes J, et al. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30:4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sarkies P, Reams C, Simpson LJ, Sale JE. Epigenetic instability due to defective replication of structured DNA. Mol Cell. 2010;40:703–713. doi: 10.1016/j.molcel.2010.11.009. The authors show that failure to properly replicate through G4 motifs affects chromatin structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodriguez R, et al. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nature Chem Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheung I, Schertzer M, Rose A, Lansdorp PM. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nature Genet. 2002;31:405–409. doi: 10.1038/ng928. Genetic experiments reveal that deficiencies in the DOG-1 helicase lead to genome instabilty at G-rich sequences in C. elegans. [DOI] [PubMed] [Google Scholar]
- 69.Kruisselbrink E, et al. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr Biol. 2008;18:900–905. doi: 10.1016/j.cub.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 70.Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. This study indicates that G4 structures cause vertebrate telomere fragility, which can be counteracted by the RTEL1 helicase. [DOI] [PubMed] [Google Scholar]
- 71.Barber LJ, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. Computational analysis revealed that G4 motifs are significantly enriched at promoters in human DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yadav VK, Abraham JK, Mani P, Kulshrestha R, Chowdhury S. QuadBase: genome-wide database of G4 DNA—occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2008;36:D381–D385. doi: 10.1093/nar/gkm781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mullen MA, et al. RNA G-quadruplexes in the model plant species Arabidopsis thaliana: prevalence and possible functional roles. Nucleic Acids Res. 2010;38:8149–8163. doi: 10.1093/nar/gkq804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nature Struct Mol Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 78.Sun D, Hurley LH. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J Med Chem. 2009;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brooks TA, Kendrick S, Hurley L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010;277:3459–3469. doi: 10.1111/j.1742-4658.2010.07759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gunaratnam M, et al. G-quadruplex compounds and cis-platin act synergistically to inhibit cancer cell growth in vitro and in vivo. Biochem Pharmacol. 2009;78:115–122. doi: 10.1016/j.bcp.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 82.Hsu ST, et al. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J Am Chem Soc. 2009;131:13399–13409. doi: 10.1021/ja904007p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Palumbo SL, Ebbinghaus SW, Hurley LH. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J Am Chem Soc. 2009;131:10878–10891. doi: 10.1021/ja902281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bejugam M, et al. Trisubstituted isoalloxazines as a new class of G-quadruplex binding ligands: small molecule regulation of c-kit oncogene expression. J Am Chem Soc. 2007;129:12926–12927. doi: 10.1021/ja075881p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marcu KB, Bossone SA, Patel AJ. myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 86.D’Cruz CM, et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nature Med. 2001;7:235–239. doi: 10.1038/84691. [DOI] [PubMed] [Google Scholar]
- 87.Strieder V, Lutz W. Regulation of N-myc expression in development and disease. Cancer Lett. 2002;180:107–119. doi: 10.1016/s0304-3835(02)00020-4. [DOI] [PubMed] [Google Scholar]
- 88.Lutz W, Leon J, Eilers M. Contributions of Myc to tumorigenesis. Biochim Biophys Acta. 2002;1602:61–71. doi: 10.1016/s0304-419x(02)00036-7. [DOI] [PubMed] [Google Scholar]
- 89.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nature Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 90.Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 91.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Han H, Langley DR, Rangan A, Hurley LH. Selective interactions of cationic porphyrins with G-quadruplex structures. J Am Chem Soc. 2001;123:8902–8913. doi: 10.1021/ja002179j. [DOI] [PubMed] [Google Scholar]
- 94.Sun D, et al. Inhibition of human telomerase by a G-quadruplex-interactive compound. J Med Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 95.Grand CL, et al. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol Cancer Ther. 2002;1:565–573. [PubMed] [Google Scholar]
- 96.Brown RV, Danford FL, Gokhale V, Hurley LH, Brooks TA. Demonstration that drug-targeted down-regulation of MYC in non-Hodgkins lymphoma is directly mediated through the promoter G-quadruplex. J Biol Chem. 2011;286:41018–41027. doi: 10.1074/jbc.M111.274720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Boddupally PV, et al. Anticancer activity and cellular repression of c-MYC by the G-quadruplex-stabilizing 11-piperazinylquindoline is not dependent on direct targeting of the G-quadruplex in the c-MYC promoter. J Med Chem. 2012;55:6076–6086. doi: 10.1021/jm300282c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gonzalez V, Guo K, Hurley L, Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J Biol Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gonzalez V, Hurley LH. The c-MYC NHE III1: function and regulation. Annu Rev Pharmacol Toxicol. 2010;50:111–129. doi: 10.1146/annurev.pharmtox.48.113006.094649. [DOI] [PubMed] [Google Scholar]
- 100.Bates PJ, Kahlon JB, Thomas SD, Trent JO, Miller DM. Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J Biol Chem. 1999;274:26369–26377. doi: 10.1074/jbc.274.37.26369. [DOI] [PubMed] [Google Scholar]
- 101.Brys A, Maizels N. LR1 regulates c-myc transcription in B-cell lymphomas. Proc Natl Acad Sci USA. 1994;91:4915–4919. doi: 10.1073/pnas.91.11.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dempsey LA, Sun H, Hanakahi LA, Maizels N. G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D, a role for G-G pairing in immunoglobulin switch recombination. J Biol Chem. 1999;274:1066–1071. doi: 10.1074/jbc.274.2.1066. [DOI] [PubMed] [Google Scholar]
- 103.Gonzalez V, Hurley LH. The C-terminus of nucleolin promotes the formation of the c-MYC G-quadruplex and inhibits c-MYC promoter activity. Biochemistry. 2010;49:9706–9714. doi: 10.1021/bi100509s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wei Q, Paterson BM. Regulation of MyoD function in the dividing myoblast. FEBS Lett. 2001;490:171–178. doi: 10.1016/s0014-5793(01)02120-2. [DOI] [PubMed] [Google Scholar]
- 105.Shklover J, Weisman-Shomer P, Yafe A, Fry M. Quadruplex structures of muscle gene promoter sequences enhance in vivo MyoD-dependent gene expression. Nucleic Acids Res. 2010;38:2369–2377. doi: 10.1093/nar/gkp1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yafe A, Shklover J, Weisman-Shomer P, Bengal E, Fry M. Differential binding of quadruplex structures of muscle-specific genes regulatory sequences by MyoD, MRF4 and myogenin. Nucleic Acids Res. 2008;36:3916–3925. doi: 10.1093/nar/gkn340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fernando H, et al. Genome-wide analysis of a G-quadruplex-specific single-chain antibody that regulates gene expression. Nucleic Acids Res. 2009;37:6716–6722. doi: 10.1093/nar/gkp740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Johnson JE, Cao K, Ryvkin P, Wang LS, Johnson FB. Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res. 2010;38:1114–1122. doi: 10.1093/nar/gkp1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wyatt JR, Davis PW, Freier SM. Kinetics of G-quartet-mediated tetramer formation. Biochemistry. 1996;35:8002–8008. doi: 10.1021/bi960124h. [DOI] [PubMed] [Google Scholar]
- 110.Mergny JL, De Cian A, Ghelab A, Sacca B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu Z, et al. Tertiary DNA structure in the single-stranded hTERT promoter fragment unfolds and refolds by parallel pathways via cooperative or sequential events. J Am Chem Soc. 2012;134:5157–5164. doi: 10.1021/ja210399h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gray RD, Chaires JB. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008;36:4191–4203. doi: 10.1093/nar/gkn379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Edmunds CE, Simpson LJ, Sale JE. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 114.Sarkies P, et al. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012;40:1485–1498. doi: 10.1093/nar/gkr868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hiratani I, Takebayashi S, Lu J, Gilbert DM. Replication timing and transcriptional control: beyond cause and effect—part II. Curr Opin Genet Dev. 2009;19:142–149. doi: 10.1016/j.gde.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Besnard E, et al. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nature Struct Mol Biol. 2012;19:837–844. doi: 10.1038/nsmb.2339. This computational study found that G4 consensus motifs are located near origins of replication in human cells. [DOI] [PubMed] [Google Scholar]
- 117.Lobachev KS, et al. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics. 1998;148:1507–1524. doi: 10.1093/genetics/148.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pan J, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Muniyappa K, Anuradha S, Byers B. Yeast meiosis-specific protein Hop1 binds to G4 DNA and promotes its formation. Mol Cell Biol. 2000;20:1361–1369. doi: 10.1128/mcb.20.4.1361-1369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Anuradha S, Muniyappa K. Meiosis-specific yeast Hop1 protein promotes synapsis of double-stranded DNA helices via the formation of guanine quartets. Nucleic Acids Res. 2004;32:2378–2385. doi: 10.1093/nar/gkh559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu Z, Gilbert W. The yeast KEM1 gene encodes a nuclease specific for G4 tetraplex DNA: implication of in vivo functions for this novel DNA structure. Cell. 1994;77:1083–1092. doi: 10.1016/0092-8674(94)90447-2. [DOI] [PubMed] [Google Scholar]
- 122.Ghosal G, Muniyappa K. Saccharomyces cerevisiae Mre11 is a high-affinity G4 DNA-binding protein and a G-rich DNA-specific endonuclease: implications for replication of telomeric DNA. Nucleic Acids Res. 2005;33:4692–4703. doi: 10.1093/nar/gki777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ghosal G, Muniyappa K. The characterization of Saccharomyces cerevisiae Mre11/Rad50/Xrs2 complex reveals that Rad50 negatively regulates Mre11 endonucleolytic but not the exonucleolytic activity. J Mol Biol. 2007;372:864–882. doi: 10.1016/j.jmb.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 124.Cahoon LA, Seifert HS. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–767. doi: 10.1126/science.1175653. The genetics in this paper provide some of the best evidence for the existence of G4 structures in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bugaut A, Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Millevoi S, Moine H, Vagner S. G-quadruplexes in RNA biology. Wiley Interdisciplinary Rev RNA. 2012;3:495–507. doi: 10.1002/wrna.1113. [DOI] [PubMed] [Google Scholar]
- 127.Svozil D, Kalina J, Omelka M, Schneider B. DNA conformations and their sequence preferences. Nucleic Acids Res. 2008;36:3690–3706. doi: 10.1093/nar/gkn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gessner RV, Frederick CA, Quigley GJ, Rich A, Wang AH. The molecular structure of the left-handed Z-DNA double helix at 1.0-Å atomic resolution. Geometry, conformation, and ionic interactions of d(CGCGCG) J Biol Chem. 1989;264:7921–7935. doi: 10.2210/pdb1dcg/pdb. [DOI] [PubMed] [Google Scholar]
- 129.Khuu P, Sandor M, DeYoung J, Ho PS. Phylogenomic analysis of the emergence of GC-rich transcription elements. Proc Natl Acad Sci USA. 2007;104:16528–16533. doi: 10.1073/pnas.0707203104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rahmouni AR, Wells RD. Stabilization of Z DNA in vivo by localized supercoiling. Science. 1989;246:358–363. doi: 10.1126/science.2678475. [DOI] [PubMed] [Google Scholar]
- 131.Ha SC, Lowenhaupt K, Rich A, Kim YG, Kim KK. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- 132.Schroth GP, Chou PJ, Ho PS. Mapping Z-DNA in the human genome. Computer-aided mapping reveals a nonrandom distribution of potential Z-DNA-forming sequences in human genes. J Biol Chem. 1992;267:11846–11855. [PubMed] [Google Scholar]
- 133.Wang G, Christensen LA, Vasquez KM. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc Natl Acad Sci USA. 2006;103:2677–2682. doi: 10.1073/pnas.0511084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhao J, Bacolla A, Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability and evolution. Cell Mol Life Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Palecek E. Local supercoil-stabilized DNA structures. Crit Rev Biochem Mol Biol. 1991;26:151–226. doi: 10.3109/10409239109081126. [DOI] [PubMed] [Google Scholar]
- 136.Pearson CE, Zorbas H, Price GB, Zannis-Hadjopoulos M. Inverted repeats, stem-loops, and cruciforms: significance for initiation of DNA replication. J Cell Biochem. 1996;63:1–22. doi: 10.1002/(SICI)1097-4644(199610)63:1%3C1::AID-JCB1%3E3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 137.van Holde K, Zlatanova J. Unusual DNA structures, chromatin and transcription. Bioessays. 1994;16:59–68. doi: 10.1002/bies.950160110. [DOI] [PubMed] [Google Scholar]
- 138.Lobachev KS, Rattray A, Narayanan V. Hairpin- and cruciform-mediated chromosome breakage: causes and consequences in eukaryotic cells. Front Biosci. 2007;12:4208–4220. doi: 10.2741/2381. [DOI] [PubMed] [Google Scholar]
- 139.Glickman BW, Ripley LS. Structural intermediates of deletion mutagenesis: a role for palindromic DNA. Proc Natl Acad Sci USA. 1984;81:512–516. doi: 10.1073/pnas.81.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Inagaki H, et al. Chromosomal instability mediated by non-B DNA: cruciform conformation and not DNA sequence is responsible for recurrent translocation in humans. Genome Res. 2009;19:191–198. doi: 10.1101/gr.079244.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kurahashi H, et al. Palindrome-mediated chromosomal translocations in humans. DNA Repair (Amst) 2006;5:1136–1145. doi: 10.1016/j.dnarep.2006.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jain A, Wang G, Vasquez KM. DNA triple helices: biological consequences and therapeutic potential. Biochimie. 2008;90:1117–1130. doi: 10.1016/j.biochi.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Manor H, Rao BS, Martin RG. Abundance and degree of dispersion of genomic d(GA)n. d(TC)n sequences. J Mol Evol. 1988;27:96–101. doi: 10.1007/BF02138367. [DOI] [PubMed] [Google Scholar]
- 144.Bacolla A, et al. Long homopurine*homopyrimidine sequences are characteristic of genes expressed in brain and the pseudoautosomal region. Nucleic Acids Res. 2006;34:2663–2675. doi: 10.1093/nar/gkl354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang X, Haber JE. Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol. 2004;2:e21. doi: 10.1371/journal.pbio.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Owen BA, et al. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nature Struct Mol Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 147.Rolfsmeier ML, Dixon MJ, Lahue RS. Mismatch repair blocks expansions of interrupted trinucleotide repeats in yeast. Mol Cell. 2000;6:1501–1507. doi: 10.1016/s1097-2765(00)00146-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.