Abstract
The “Berlin Patient,” who maintains suppressed levels of HIV viremia in the absence of antiretroviral therapy, continues to be a standard bearer in HIV eradication research. However, the unique circumstances surrounding his functional cure are not applicable to most HIV+ patients. To achieve a functional or sterilizing cure in a greater number of infected individuals worldwide, combinatorial treatments, targeting multiple stages of the viral life cycle, will be essential. Several anti-HIV gene therapy approaches have recently been explored, including disruption of the CCR5 and CXCR4 coreceptor loci in CD4+ T-cells and CD34+ hematopoietic stem cells. However, less is known about the efficacy of these strategies in patients and more relevant HIV model systems such as nonhuman primates. Combinatorial approaches, including genetic disruption of integrated provirus, functional enhancement of endogenous restriction factors, and/or the use of pharmacological adjuvants, could amplify the anti-HIV effects of CCR5/CXCR4 gene disruption. Importantly, delivering gene disruption molecules to genetic sites of interest will likely require optimization on a cell type-by-cell type basis. In this review, we highlight the most promising gene therapy approaches to combat HIV infection, methods to deliver these therapies to hematopoietic cells, and emphasize the need to target viral replication pre- and post-entry in order to mount a suitably robust defense against spreading infection.
Introduction
As the number of HIV-positive individuals approaches 40 million worldwide, a new drive to cure HIV infection has gained ground. Although highly active antiretroviral therapy (HAART) has drastically improved the odds of survival in infected patients, research over the past decade has made it clear that HAART is not a curative therapy.1–4 Furthermore, less than 5% of the HIV-positive population in many developing countries receives HAART (UNAIDS World AIDS Day Report, 2011). Even in the first world, where coverage is significantly higher, recent studies suggest that long-term HAART therapy is associated with a number of chronic diseases.5–8 In short, although HAART has significantly decreased HIV-related mortality worldwide, a cure for HIV infection remains a highly desirable, yet elusive goal.
Substantial research over the past 5 years has focused on the in vitro and in vivo modeling of the one human patient who has potentially been cured of HIV: Timothy Brown, better known as the Berlin Patient. In 2007, during treatment for relapsed acute myeloid leukemia, this HIV+ patient received an allogeneic hematopoietic stem cell transplant (HCT) from a donor homozygous for the Δ32 null mutation in CCR5;9 these cells should be resistant to infection by CCR5-tropic HIV-1. Consistent with this prediction, the Berlin patient displays no detectable viremia to date, despite cessation of HAART.10,11 Although the mechanism(s) behind the eradication of HIV in this patient are unclear, multiple factors may have contributed. In addition to the CCR5-null donor cells themselves, the preparative regimens administered prior to HCT likely ablated a proportion of latently-infected host CD4+ cells, and Grade I graft-versus-host disease (GVHD) may have further depleted the latent reservoir.12,13 The most likely explanation is that a combination of these factors each contributed to the long-term, HAART-independent control of HIV-1 in this patient.14
The Berlin Patient has served as the groundwork for an expanded field of stem cell gene therapy approaches aimed at preventing HIV replication. Many, including targeting CCR5 and other host genes, whose modification may lead to the generation of HIV-resistant progenitor cells, have been reviewed elsewhere.15–18 Recently, focus has shifted to how combinatorial approaches could increase resistance to infection. In this review, we will discuss some of the most promising stem cell-based anti-HIV therapies, outline strategies for delivering targeting molecules to hematopoietic cells, and highlight steps throughout the viral life cycle whose synergistic targeting may lead to HAART-independent control of HIV infection in vivo.
The HIV Replication Cycle Offers Multiple Potential Targets For Genetic Interruption of Infection
The viral envelope (env) protein displayed by mature HIV-1 virions interacts with the cell surface glycoprotein CD4, which is expressed on helper T cells, dendritic cells, and monocyte/macrophages (Figure 1a).19,20 This interaction is stabilized by env binding to one of two coreceptor molecules, CCR5 or CXCR4, which in turn triggers viral membrane fusion to the host plasma membrane, and release of the encapsidated, dimeric RNA genome into the cytosol. A subset of hematopoietic precursor cells may also be susceptible to low levels of HIV entry, although this point remains controversial.21–24 After uncoating of the encapsidated viral RNA in the infected cell, a bound molecule of viral reverse transcriptase converts the RNA to DNA, which is imported into the nucleus, and integrates into the host genome. Coordinated transcription and translation lead to production of new viral RNA and proteins, which complete assembly at the plasma membrane and bud a new virus.25
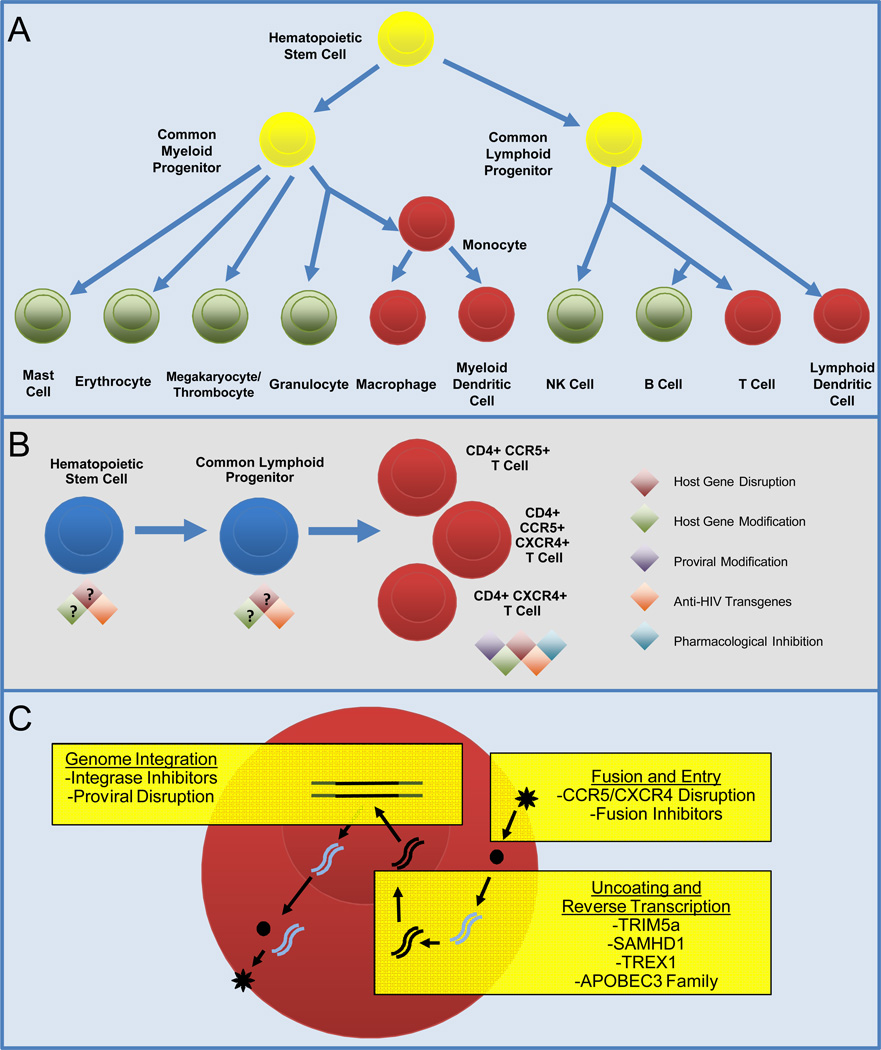
Figure 1. Applicable Stages and Strategies for Gene Therapy-Based Antagonism of HIV Replication.
A) Simplified diagram of hematopoiesis, highlighting myeloid and lymphoid lineages that are susceptible to HIV infection (red), or are refractory to infection (green). Yellow: HIV infection of hematopoietic progenitors remains controversial. B) Schematic of T cell maturation, indicating applicable anti-HIV therapies at each stage of differentiation. Disruption or modification of some host genes may preferably be performed in more differentiated cells. C): Three broad access points for inhibition of HIV infection and replication. HIV entry can be blocked by disrupting HIV coreceptor genes, and/or expression of fusion inhibitors. Genetic modification of TRIM5α, SAMHD1, TREX1, or APOBEC proteins may increase antagonism of viral uncoating and reverse transcription. Pharmacological inhibition of HIV integrase, or targeted disruption of integrated provirus, eliminate or prevent establishment of the latent viral reservoir.
Inhibition of viral entry is one of the best-studied means of preventing viral replication. Whether through modification of the HIV coreceptors CCR5 or CXCR416,26 or blocking membrane fusion through dominant negative peptides and small molecules,27 inhibition of HIV entry prevents viral replication by sequestering viral particles outside of the cell. Importantly, multiple subsequent stages in the viral life cycle may also be targeted, including capsid uncoating, proviral integration, and targeted ablation of the integrated provirus itself (Figure 1C).28 Although these latter stages have received less attention in HIV eradication studies, their combination with entry-based approaches may prove essential in the development of curative therapies. We will discuss the application of these strategies in hematopoietic stem cells, which may allow for the protection of multiple hematopoietic lineages, and in differentiated hematopoietic cells such as CD4+ T-cells, which are directly impacted by HIV infection (Figure 1A–1B).
CCR5 Disruption and Other anti-HIV Entry Therapies
Inhibition of CCR5 is among the best-characterized means of blocking HIV-1 entry. This strategy is particularly advantageous because loss of CCR5 expression is not associated with any significant pathologies;29,30 in fact, the CCR5Δ32 null mutation is stably inherited in a sizeable population in Europe and western Asia.31–33 These findings indicate that the targeted disruption of CCR5 should protect an individual from infection with CCR5-tropic virus, without significantly impacting other cellular functions.
Genetic disruption of CCR5 has been documented in human CD34+ hematopoietic stem cells as well as CD4+ T-helper cells. Perez and colleagues demonstrated robust disruption of CCR5 in CD4+ cells, engraftment in a NOG mouse model of HIV-1 infection, and enrichment during HIV-1 challenge. These autologous, HIV-resistant CD4+ T cells are currently being tested in Phase I clinical trials as a novel antiviral therapeutic.34,35 Holt and colleagues performed analogous experiments in human cord blood-derived CD34+ stem cells. These cells engrafted and repopulated hematopoietic niches in all tissues tested in NSG mice, and were enriched following HIV-1 challenge.36 More recently, CD34+ cells transduced with a lentivirus expressing CCR5 shRNA, a TAR element decoy, and a rhesus macaque allele of TRIM5α that restricts HIV-1 were also shown to resist infection and confer partial protection against HIV in vitro and in humanized mice.37,38 Notably, few studies to date have assessed the efficacy of CCR5-targeted CD34+ cells in more relevant nonhuman primate models or in clinical trials. Nonetheless, experiments in mouse models suggest that engineering HIV-resistant hematopoietic cells via targeting of CCR5 is a feasible strategy, and can be achieved in multiple relevant cell types.
One significant challenge inherent in translating genetic disruption of CCR5 to a larger animal model or clinical trial is the ability to produce gene-targeted hematopoietic cells in relevant quantities, and with sufficient levels of CCR5 disruption. For example, Holt and colleagues demonstrated that CD34+ cells with biallelic CCR5 disruption frequencies as low as 5–7% were still capable of resisting HIV infection in murine experiments, but also noted that DNA nucleofection, which they used to deliver CCR5-disrupting molecules into CD34+ cells, was associated with relatively high toxicity.36 Therefore, it is critical to identify the combination of cell culture, gene delivery, and CCR5 disruption conditions that maximizes cell number, viability, function, and CCR5 loss of function.
General methods to disrupt a targeted gene of interest, such as CCR5, have been reviewed elsewhere.39–41 Briefly, the three best characterized methods, zinc finger nucleases (ZFNs), homing endonucleases (HEs) and TAL effector nucleases (TALENs) each work in an analogous fashion. Sequence specific DNA binding domain(s) target the locus of interest, and a DNA endonuclease domain creates a double strand break (DSB), which is either repaired by non-homologous end joining (NHEJ) or by homologous recombination (HR).42 HR allows for the incorporation of reporter genes or preferential alleles at a targeted locus; so far, it has proven more successful in cell lines than in primary cells.43,44 Perez et al and Holt et al both exploited the NHEJ pathway to disrupt CCR5, wherein random nucleotide insertions or deletions during DSB repair introduce frameshift mutations, often generating a null allele.35,36 While NHEJ is useful in the disruption of non-essential genes like CCR5, less-efficient HR pathways must be invoked at target genes for which random mutagenesis could lead to secondary pathologies.
In addition to robust disruption of the targeted locus, it is imperative that any method of targeted disruption be highly specific. A greater number of aberrant cleavage events raises the possibility that an important off-target gene may be disrupted, or that DSB-induced apoptosis may be triggered in the targeted cell.45,46 Rational design and thorough vetting using deep sequencing have significantly increased the specificity of gene disruption molecules;47,48 DSB-induced apoptosis may be a greater obstacle in some primary cell types. Whereas transformed and immortalized cell lines adapt to higher levels of genotoxic events like DSBs, repeated rounds of DSB induction and DNA repair in primary cells may more rapidly signal to cell death pathways.49,50
As mentioned above, only a fraction of cells will acquire mutations at both alleles of CCR5 following genetic targeting. By combining this gene therapy approach with pharmacological approaches, antagonism of HIV entry may be further enhanced. For example, peptides such as C46 and T20/enfuvirtide act as dominant negatives to block membrane fusion (refs.51–53 and Younan et al. submitted). CCR5-binding small molecules like INCB9471, Vicriviroc, Aplaviroc, and most notably Mariviroc, block association with env and are predicted to phenocopy CCR5 genetic disruption.54–56 Antibody-based therapies also block env interactions with the cell surface: TNX-355,57, which binds CD4, PRO 140,58 which binds CCR5, and b12,59 the best-characterized HIV-1 neutralizing antibody. Finally, synthetic (BMS-488043, DCM205) and naturally occurring (Griffithsin, Epigallocatechin gallate) small molecules also disrupt the interaction between HIV-1 and CD4+ cells.60–63 Combinations of the aforementioned anti-HIV entry treatments have demonstrated synergistic antagonism of HIV entry.64 Used as an adjuvant with CCR5 gene therapy, these drugs may offer further resistance to CCR5-tropic virus.
Since CD4 plays an essential role in adaptive immune responses,65,66 genetic targeting of the CD4 gene is not a feasible approach. However, therapies that block interaction between HIV-1 and CD-4 without impinging on CD4 function, such as the non-immunosuppressive TNX-355 antibody, may prove useful in blocking viral entry. In addition, transient inhibition of CD4 has proven useful in a number of autoimmune diseases,67,68 and could similarly be used as a short-term adjuvant, for example during activation and clearance of latently infected CD4+ T-cells.
A major concern with all of the above anti-CCR5 therapies is the possible selection for tropism-switching viruses, i.e. viruses that evolve to use CXCR4 instead of CCR5 as a coreceptor at the surface of CD4+ cells. One solution would be to target both CCR5 and CXCR4, since CXCR4 disruption has been demonstrated to reduce HIV infection in a humanized mouse model, similar to CCR5 disruption.69,70 Unlike CCR5, no stable CXCR4 null mutations exist in human populations, and CXCR4 null mice die during early embryonic development due to multiple anomalies in hematopoietic, vascular, and neuronal development.71 T-cell-specific CXCR4 knockout mice, however, are viable and lack appreciable T-cell defects,72 and Wilen and colleagues have demonstrated that CCR5/CXCR4 doubly-disrupted T cells are viable in vitro.69 In sum, CCR5/CXCR4 disruption is likely intractable in hematopoietic stem cells, but may represent a viable combinatorial therapy in CD4+ T-cells (Table 1).
Table 1.
Summary of Anti-HIV Gene Therapy Approaches and their Applicability in Hematopoietic Stem Cells and Differentiated Hematopoietic Cells
| Strategy | Stem Cells | Mature Cells† |
References |
|---|---|---|---|
| CCR5 disruption | Yes | Yes | 12,35,36 |
| CXCR4 disruption | No | Yes | 69,70 |
| CCR5/CXCR4 double disruption | No | Yes | 69 |
| Restriction factors: | |||
| Transgenic expression | Unclear* | Yes | 37,38 |
| Gene modification at endogenous locus | Unclear* | Yes | 76,77,83,114 |
| Disruption of HIV provirus‡ | Yes‡ | Yes | 98,99 |
Transgenic or targeted expression of enhanced restriction factors in hematopoietic precursor cells may have unforeseen effects on hematopoietic differentiation or function of differentiated hematopoietic cells.
Macrophage, DC, and T-Cell subsets; see Figure 1A.
Currently, detection of proviral DNA in hematopoietic precursor cells remains controversial.
Inhibition of Viral Uncoating and Integration
Cellular restriction factors target and antagonize HIV replication at multiple steps during the viral life cycle, in particular, immediately after the virus enters the host cell (reviewed in73,74). Of the five best-characterized restriction factors to date (TRIM5α, TREX1, SAMHD1, the APOBEC3 family, and BST-2), only BST-2, which antagonizes viral release, does not target viral replication between entry and proviral integration.75 Furthermore, the vast majority of HAART therapies include at least one component targeting these stages, either inhibitors of HIV reverse transcriptase or HIV integrase.28 Collectively, these observations suggest that the post-entry uncoating, reverse transcription, and genome integration of HIV are highly amenable to antagonism (Figure 1C).
In the context of curative strategies for HIV, post-entry/pre-integration is less emphasized relative to inhibition of entry itself. However, several recent publications demonstrate that viral eradication may be more easily achieved by antagonizing multiple early stages of the viral life cycle, from viral entry to integration. For example, Walker and colleagues transduced human hematopoietic stem cells with a triple lentiviral vector expressing a CCR5 shRNA to antagonize viral entry, a rhesus macaque allele of TRIM5α to antagonize viral uncoating, and a viral TAR decoy to down-regulate reverse transcription of viral RNA.37 This triple protection led to robust resistance against HIV-1 in a humanized mouse model.
The vast body of literature describing the competition between the restriction factor APOBEC3G (A3G) and the HIV accessory protein Vif has recently been modeled in silico, and should prove useful to anti-HIV gene therapy approaches.76 Utilizing algorithms that describe the in vitro interaction between A3G and Vif, the authors predicted that an A3G mutant that was unable to bind Vif was relatively more resistant to infection than ubiquitination-resistant A3G mutants, or cells treated with an antibody which blocks the interaction of Vif with A3G. However, the authors’ model carried the caveat that these therapies must be delivered to nearly 100% of cells to show an effect. These computational studies will be essential in order to translate in vitro data to relevance in large animal models and clinical trials; similar approaches could be used to rationally construct an ideal gene modification at any anti-HIV restriction factor of interest. For example, a single amino acid substitution in the TRIM5α B30.2/SPRY domain could restore its ability to bind viral capsid protein and disrupt post-entry viral uncoating.77 Introduction of the D128K mutation into APOBEC3G should block antagonism by Vif and restore A3G’s cytidine deaminase activity on viral RNA.78
Although these optimized restriction factors can be delivered to hematopoietic cells as viral transgenes, the risks associated with transgenic viral delivery preclude this strategy.16,79 In addition, both TRIM5α and APOBEC3G function as multimers.80,81 Hence, endogenous TRIM5α or APOBEC3G could dilute the enhanced function of exogenous TRIM5α P332R or APOBEC3G D128K, respectively, in a mixed oligomer. To avoid these risks, gene disruption technology could be combined with homologous recombination to target restriction-enhancing mutations to the endogenous locus. The double strand break generated by targeted endonucleases like HEs, ZFNs, and TALENs could be utilized for repair by homologous recombination in the presence of a donor cassette carrying the modified allele of the targeted gene. Recent work demonstrates that this engineering can be performed with non-integrating lentiviruses and cell electroporation.82,83 This strategy would allow a gene modification to be introduced at a locus of interest while avoiding all risks associated with viral insertion sites and transgenic expression.
Whether these optimized restriction factors are expressed as transgenes or targeted to the endogenous locus, it is important to consider their function in non-pathogenic contexts. APOBEC proteins edit a number of cellular mRNAs in addition to viral RNAs,84,85 while TRIM5 proteins activate NF-kB and AP-1-dependent transcription in a virus-independent manner.86,87 Therefore, it will be essential to confirm that expression of modified alleles, especially in hematopoietic precursor cells, does not alter cellular differentiation or function in differentiated hematopoietic cell types (Table 1).88
In sum, using targeted endonucleases to directly modify a restriction factor and enhance its antiviral activity is a key and emerging tool in combinatorial anti-HIV gene therapy, and could be used in tandem with analogous gene disruption strategies, for instance at the CCR5 locus.
Disruption of HIV Provirus
Until recently, HIV eradication studies have centered on two broad strategies: prevent infection prior to proviral integration, as described above, or target latently infected cells for destruction. Although only one out of 1 million CD4+ T cells in HAART-treated patients harbors latent HIV-1 provirus,89 this viral reservoir is quickly reactivated following cessation of HAART.90,91 Therefore, a substantial effort has focused on targeting latently infected cells for viral reactivation.92–94 These cells should then be destroyed by the host immune response, although additional strategies to completely clear these virus-producing cells will likely be needed.95 Since this approach carries the potential to eradicate some, if not all, of the latent HIV reservoir, it is a major focus of HIV curative therapies, and several strategies are currently being evaluated in clinicial trials.96 An important presumption in these trials is that infected cells will be destroyed at a greater rate than reactivated virus is able to spread.97 Ideally, latently infected, mature hematopoietic cells would be targeted without the risks associated with viral reactivation.
Recently, we have genetically disrupted integrated provirus in infected cells using methods analogous to genetic disruption of CCR5.98 Using homing endonucleases modified to target conserved sequences within HIV-1, such an approach is feasible, and could be designed to target multiple proviral sites, thus avoiding the problem of viral escape. Importantly, this is the only strategy to date that targets latently infected cells and does not require viral reactivation.99 It will be interesting to evaluate whether proviral disruption is enhanced in reactivated cells, where chromatin structure at proviral integration sites is more open and accessible.100 More broadly, combining viral reactivation methods with strategies to prevent viral replication, like CCR5 disruption, could enhance depletion of the viral reservoir while lessening the risk of spreading infection.
Delivery of Therapeutic Molecules
Most HIV eradication studies to date have used humanized mouse models to demonstrate efficacy of approaches such as CCR5 disruption in vivo; modeling these strategies in relevant nonhuman primate (NHP) models of HIV infection is the next logical step. Gene disruption studies in NHPs will require redesigned targeted endonucleases that recognize NHP genes of interest. In addition, our preliminary data suggest that gene delivery strategies must be specifically tailored for human versus primate cells: the strategies that work best in human CD34+ and CD4+ cells are usually suboptimal in NHP cells (our unpublished data). Therefore, it is important to discuss the various gene delivery options available in the two cell types most relevant to HIV-1 infection: CD34+ hematopoietic stem cells and CD4+ T Helper cells.
As discussed above, integrating retroviruses offer sustained, robust expression of anti-HIV transgenes, including exogenous restriction factors37 and peptides (52,53 and Younan et al submitted). For therapeutic use, the long tandem repeats in these viruses must be extensively engineered in order to minimize their promoter activity and consequent oncogenic risks.16 Furthermore, identification of the ideal transducing retrovirus is often dependent on cell type and species, and must consider the best retroviral subtype and pseudotype for optimal gene delivery.79 An important development over the past several years has been the advent of non-integrating retroviruses that are able to express transgenes of interest, but are not able to integrate into the host genome.83,101 In addition to the advantage of avoiding insertional mutagenesis, these vectors also facilitate robust expression of genes of interest over shorter time frames. This is ideal for the delivery of targeting endonucleases, whose longer-term expression may lead to off-target cleavage events. Importantly, non-integrating lentiviruses may still integrate into host genomes at low frequencies, although the relative incidence of such events remains controversial.102,103
Adenoviral delivery holds an advantage over lentiviral delivery in that adenoviruses are intrinsically non-integrating.104,105 Adenovirus has proven especially useful in the delivery of CCR5- and CXCR4-modifying enzymes into human and macaque CD4 cells.35,69,70 Two cautionary notes must be made when considering adenoviral delivery-based gene therapy approaches. First, the production of adenoviral particles is among the most time-consuming of any vector system, taking several weeks as compared to several days for retrovirus production.106,107 Second, the well-documented failure of the HIV-1 STEP trial has raised questions for the clinical use of adenoviral-mediated gene delivery: since adenoviruses activate a subset of CD4+ cells, and activated CD4+ cells are at highest risk for infection by HIV, adenoviral delivery methods may actually increase short-term susceptibility to HIV infection.108,109 However, use of adenoviral vectors to target genes of interest in primary cells ex vivo is likely to carry significantly less risk than their use in vivo in vaccine trials such as STEP.
Adeno-associated viruses, like adenoviruses, do not integrate into the host genome, but are drastically less time-consuming to produce. AAV serotype 2-based vectors are the best-characterized for transduction of human hematopoietic cells, although AAV2 immunogenicity is still under debate.110–113 Further work demonstrates that AAV vectors may be ideally suited for the introduction of donor cassettes for homologous recombination following gene disruption.114 In sum, AAV vectors offer a promising mode of delivery for both anti-HIV transgenes and gene modification templates, but require extensive further testing in most model systems. In nonhuman primate models of HIV, for instance, the use of AAV-mediated gene delivery remains in early stages, and may require re-optimization relative to findings in human cells.115
Two non-viral gene delivery strategies have emerged in anti-HIV therapeutics. Cell electroporation has gained popularity as various devices and electrochemical programs have been optimized to maximize delivery of DNA and RNA cargoes at minimal cost to cell viability. These parameters have led to the successful transduction of CCR5-specific ZFNs into human cord blood CD34+ cells, which were able to engraft into humanized mice and expand following HIV-1 challenge.36 As discussed above, cell electroporation studies will likely need to be designed on a species- and cell type-specific basis to optimize cell viability and gene delivery.
Nucleic acid aptamers and dendrimers116,117 have been designed to target both CD4 and HIV env, and represent another non-viral means of anti-HIV gene delivery. These antibody-like molecules can be conjugated to antiviral or pro-apoptotic mRNAs or shRNAs, which are endocytosed after binding to their specific cell surface epitope.118 An important consideration for both mRNA electroporation and aptamer/dendrimer-based gene delivery is the stability of these molecules in vivo. While DNA delivery is more stable, it is also more toxic, especially in primary cells;119 synthesis of mRNA cargoes using pseudo-uridine and 5-methyl cytosine imparts stability comparable to DNA, without associated toxicity.119,120
Conclusions and Future Prospects
The Berlin patient has catalyzed the field of stem cell-based anti-HIV therapy. However, to make these approaches more applicable to a larger number of patients will most likely require the introduction of additional HIV resistance factors, concurrent with stem cell-based treatments. Here we reviewed the breadth of stem cell-based gene therapy approaches currently in development, additional anti-HIV treatments that could be used in combination, and potential modes of delivery for these therapies. While the majority of work centers on blocking viral entry in uninfected cells and destroying latently infected cells, other emerging strategies have shown great promise as well. In combination, these methods could potentially minimize HIV disease burden to levels associated with disease eradication. Alternatively, these methods may result in a functional cure, wherein a smaller proportion of cells could still harbor HIV provirus, and extremely low levels of viremia may still be detectable, yet patients would remain aviremic in the absence of HAART.
We have sought to emphasize the need to target the viral life cycle at multiple steps in order to eradicate HIV beyond the need for HAART treatment. It is likely that focus on any one step will result in escape mutants which could in fact increase viral burden: anti-CCR5 therapies could induce CCR5-CXCR4 tropism switching, compensatory viral mutations may arise in response to modification of viral restriction factors, and resistance mutations may arise in response to antibody-based inhibitors of viral entry. HAART advanced markedly as drugs evolved and were administered in rational combinations, targeting multiple stages of viral replication. Likewise, the integration of multiple molecular and genetic blocks to HIV infection may further extend our control of this disease, to levels where HAART cessation may be feasible. The recent developments in stem cell-based anti-HIV therapies, the ability to genetically and pharmacologically target multiple stages of the viral life cycle, and the ability to safely deliver these therapies to infected cells, promise new curative treatments that should prove applicable to HIV patients worldwide.
Acknowledgments
The authors would like to acknowledge the impact of NIH-supported Martin Delaney Collaboratory grant U19 AI 096111 to H.P.K and K.R.J. This effort was supported in part by R01 AI80326 and R01 HL84345 (to H.P.K.), and Bill and Melinda Gates Foundation Grand Challenges Explorations Phase I award 51763 and Phase II award OPP1018811 (to K.R.J.). H.P.K. is a Markey Molecular Medicine Investigator and also supported by the Jose´ Carreras/E.Donnall Thomas Endowed Chair for Cancer Research.
Reference List as of 11-13-2012
- 1.Camacho R, Teofilo E. Antiretroviral therapy in treatment-naive patients with HIV infection (Review) Current Opinion in HIV & AIDS. 2011;6(Suppl 1):S3–S11. doi: 10.1097/01.COH.0000410239.88517.00. [DOI] [PubMed] [Google Scholar]
- 2.May MT, Ingle SM. Life expectancy of HIV-positive adults: a review (Review) Sexual Health. 2011;8:526–533. doi: 10.1071/SH11046. [DOI] [PubMed] [Google Scholar]
- 3.Shen L, Siliciano RF. Viral reservoirs, residual viremia, and the potential of highly active antiretroviral therapy to eradicate HIV infection (Review) Journal of Allergy & Clinical Immunology. 2008;122:22–28. doi: 10.1016/j.jaci.2008.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams BG, Lima V, Gouws E. Modelling the impact of antiretroviral therapy on the epidemic of HIV (Review) Current HIV Research. 2011;9:367–382. doi: 10.2174/157016211798038533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blas-Garcia A, Apostolova N, Esplugues JV. Oxidative stress and mitochondrial impairment after treatment with anti-HIV drugs: clinical implications (Review) Current Pharmaceutical Design. 2011;17:4076–4086. doi: 10.2174/138161211798764951. [DOI] [PubMed] [Google Scholar]
- 6.Fisher SD, Kanda BS, Miller TL, Lipshultz SE. Cardiovascular disease and therapeutic drug-related cardiovascular consequences in HIV-infected patients (Review) American Journal of Cardiovascular Drugs. 2011;11:383–394. doi: 10.2165/11594590-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 7.Mothobi NZ, Brew BJ. Neurocognitive dysfunction in the highly active antiretroviral therapy era (Review) Curr Opin Inf Dis. 2012;25:4–9. doi: 10.1097/QCO.0b013e32834ef586. [DOI] [PubMed] [Google Scholar]
- 8.Palios J, Kadoglou NP, Lampropoulos S. The pathophysiology of HIV−/HAART−related metabolic syndrome leading to cardiovascular disorders: the emerging role of adipokines (Review) Experimental Diabetes Research. 2012;2012:103063. doi: 10.1155/2012/103063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 10.Allers K, Hutter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, et al. Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood. 2011;117:2791–2799. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 11.Hutter G, Thiel E. Allogeneic transplantation of CCR5-deficient progenitor cells in a patient with HIV infection: an update after 3 years and the search for patient no. 2. AIDS. 2011;25:273–274. doi: 10.1097/QAD.0b013e328340fe28. [DOI] [PubMed] [Google Scholar]
- 12.Hutter G, Ganepola S. Eradication of HIV by transplantation of CCR5-deficient hematopoietic stem cells. The Scientific World Journal. 2011;11:1068–1076. doi: 10.1100/tsw.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamp C, Wolf T, Bravo IG, Kraus B, Krause B, Neumann B, et al. Decreased HIV diversity after allogeneic stem cell transplantation of an HIV-1 infected patient: a case report. Virology Journal. 2010;7:55. doi: 10.1186/1743-422X-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutter G, Zaia JA. Allogeneic haematopoietic stem cell transplantation in patients with human immunodeficiency virus: the experiences of more than 25 years (Review) Clin Exp Immunol. 2011;163:284–295. doi: 10.1111/j.1365-2249.2010.04312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durand C, Ambinder R, Blankson J, Forman S. HIV-1 and hematopoietic stem cell transplantation (Review) Biol Blood Marrow Transplant. 2012;18:S172–S176. doi: 10.1016/j.bbmt.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiem H-P, Jerome KR, Deeks SG, McCune JM. Hematopoietic-stem-cell-based gene therapy for HIV disease [Review] Cell Stem Cell. 2012;10:137–147. doi: 10.1016/j.stem.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai Y. CCR5-targeted hematopoietic stem cell gene approaches for HIV disease: current progress and future prospects. Current Stem Cell Research & Therapy. 2012;7:310–317. doi: 10.2174/157488812800793108. [DOI] [PubMed] [Google Scholar]
- 18.Scherer LJ, Rossi JJ. Ex vivo gene therapy for HIV-1 treatment (Review) Hum Mol Genet. 2011;20:R100–R107. doi: 10.1093/hmg/ddr160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arrildt KT, Joseph SB, Swanstrom R. The HIV-1 env protein: a coat of many colors (Review) Current HIV/AIDS Reports. 2012;9:52–63. doi: 10.1007/s11904-011-0107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilen CB, Tilton JC, Doms RW. Molecular mechanisms of HIV entry (Review) Advances in Experimental Medicine & Biology. 2012;726:223–242. doi: 10.1007/978-1-4614-0980-9_10. [DOI] [PubMed] [Google Scholar]
- 21.Durand CM, Ghiaur G, Siliciano JD, Rabi SA, Eisele EE, Salgado M, et al. HIV-1 DNA is detected in bone marrow populations containing CD4+ T cells but is not found in purified CD34+ hematopoietic progenitor cells in most patients on antiretroviral therapy. J Infect Dis. 2012;205:1014–1018. doi: 10.1093/infdis/jir884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Josefsson L, Eriksson S, Sinclair E, Ho T, Killian M, Epling L, et al. Hematopoietic precursor cells isolated from patients on long-term suppressive HIV therapy did not contain HIV-1 DNA. J Infect Dis. 2012;206:28–34. doi: 10.1093/infdis/jis301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNamara LA, Collins KL. Hematopoietic stem/precursor cells as HIV reservoirs (Review) Current Opinion in HIV & AIDS. 2011;6:43–48. doi: 10.1097/COH.0b013e32834086b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNamara LA, Ganesh JA, Collins KL. Latent HIV-1 infection occurs in multiple subsets of hematopoietic progenitor cells and is reversed by NF-kappa beta activation. J Virol. 2012 doi: 10.1128/JVI.00895-12. [Epub ahead of print 2012 Jun 20]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedrich BM, Dziuba N, Li G, Endsley MA, Murray JL, Ferguson MR. Host factors mediating HIV-1 replication (Review) Virus Research. 2011;161:101–114. doi: 10.1016/j.virusres.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Loftin LM, Kienzle M, Yi Y, Collman RG. R5X4 HIV-1 coreceptor use in primary target cells: implications for coreceptor entry blocking strategies (Review) Journal of Translational Medicine. 2011;9(Suppl 1):S3. doi: 10.1186/1479-5876-9-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Didigu CA, Doms RW. Novel approaches to inhibit HIV entry (Review) Viruses. 2012;4:309–324. doi: 10.3390/v4020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeier MD, Nachega JB. Targeting HIV: past, present and future (Review) Infectious Disorders - Drug Targets. 2011;11:98–114. doi: 10.2174/187152611795589690. [DOI] [PubMed] [Google Scholar]
- 29.Melum E, Karlsen TH, Broome U, Thorsby E, Schrumpf E, Boberg KM, et al. The 32-base pair deletion of the chemokine receptor 5 gene (CCR5-Delta32) is not associated with primary sclerosing cholangitis in 363 Scandinavian patients. Tissue Antigens. 2006;68:78–81. doi: 10.1111/j.1399-0039.2006.00604.x. [Erratum appears in Tissue Antigens. 2006 Aug;68(2)192] [DOI] [PubMed] [Google Scholar]
- 30.Muntinghe FL, Carrero JJ, Navis G, Stenvinkel P. TNF-alpha levels are not increased in inflamed patients carrying the CCR5 deletion 32. Cytokine. 2011;53:16–18. doi: 10.1016/j.cyto.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Kamkamidze G, Capoulade-Metay C, Butsashvili M, Dudoit Y, Chubinishvili O, Debre P, et al. 32-nucleotide deletion, associated with defence against hiv/aids, is a predominant mutation of CCR5 gene in the population of Georgia. Georgian Medical News. 2005:74–79. [PubMed] [Google Scholar]
- 32.Novembre J, Galvani AP, Slatkin M. The geographic spread of the CCR5 Delta32 HIV-resistance allele. PLoS Biology. 2005;3:e339. doi: 10.1371/journal.pbio.0030339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poljak M, Maver PJ, Seme K. Prevalence of CCR5 gene 32-basepair deletion in populations of Slavic origin. Croatian Medical Journal. 2006;47:348–349. [PMC free article] [PubMed] [Google Scholar]
- 34.Cannon P, June C. Chemokine receptor 5 knockout strategies (Review) Current Opinion in HIV & AIDS. 2011;6:74–79. doi: 10.1097/COH.0b013e32834122d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker JE, Chen RX, McGee J, Nacey C, Pollard RB, Abedi M, et al. Generation of an HIV-1-resistant immune system with CD34(+) hematopoietic stem cells transduced with a triple-combination anti-HIV lentiviral vector. J Virol. 2012;86:5719–5729. doi: 10.1128/JVI.06300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson JS, Javien J, Nolta JA, Bauer G. Preintegration HIV-1 inhibition by a combination lentiviral vector containing a chimeric TRIM5 alpha protein, a CCR5 shRNA, and a TAR decoy. Mol Ther. 2009;17:2103–2114. doi: 10.1038/mt.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carroll D. Genome engineering with zinc-finger nucleases (Review) Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scholze H, Boch J. TAL effectors are remote controls for gene activation (Review) Current Opinion in Microbiology. 2011;14:47–53. doi: 10.1016/j.mib.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Stoddard BL. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification (Review) Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grabarz A, Barascu A, Guirouilh-Barbat J, Lopez BS. Initiation of DNA double strand break repair: signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining. American Journal of Cancer Research. 2012;2:249–268. [PMC free article] [PubMed] [Google Scholar]
- 43.Munoz IG, Prieto J, Subramanian S, Coloma J, Redondo P, Villate M, et al. Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus. Nucleic Acids Res. 2011;39:729–743. doi: 10.1093/nar/gkq801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Overlack N, Goldmann T, Wolfrum U, Nagel-Wolfrum K. Gene repair of an usher syndrome causing mutation by zinc-finger nuclease mediated homologous recombination. Investigative Ophthalmology & Visual Science. 2012;53:4140–4146. doi: 10.1167/iovs.12-9812. [DOI] [PubMed] [Google Scholar]
- 45.Ghardi M, Moreels M, Chatelain B, Chatelain C, Baatout S. Radiation-induced double strand breaks and subsequent apoptotic DNA fragmentation in human peripheral blood mononuclear cells. Int J Mol Med. 2012;29:769–780. doi: 10.3892/ijmm.2012.907. [DOI] [PubMed] [Google Scholar]
- 46.Mittelman D, Moye C, Morton J, Sykoudis K, Lin Y, Carroll D, et al. Zinc-finger directed double-strand breaks within CAG repeat tracts promote repeat instability in human cells. Proc Natl Acad Sci USA. 2009;106:9607–9612. doi: 10.1073/pnas.0902420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrmann F, Garriga-Canut M, Baumstark R, Fajardo-Sanchez E, Cotterell J, Minoche A, et al. p53 Gene repair with zinc finger nucleases optimised by yeast 1-hybrid and validated by Solexa sequencing. PLoS ONE. 2011;6:e20913. doi: 10.1371/journal.pone.0020913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reyon D, Kirkpatrick JR, Sander JD, Zhang F, Voytas DF, Joung JK, et al. ZFNGenome: a comprehensive resource for locating zinc finger nuclease target sites in model organisms. BMC Genomics. 2011;12:83. doi: 10.1186/1471-2164-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cassidy LD, Venkitaraman AR. Genome instability mechanisms and the structure of cancer genomes (Review) Curr Opin Genet Dev. 2012;22:10–13. doi: 10.1016/j.gde.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 50.Cunderliková B. Issues to be considered when studying cancer in vitro. Crit Rev Oncol Hematol. 2012 doi: 10.1016/j.critrevonc.2012.06.007. [Epub ahead of print 2012 Jul 21]. [DOI] [PubMed] [Google Scholar]
- 51.Qiu S, Yi H, Hu J, Cao Z, Wu Y, Li W. The binding mode of fusion inhibitor T20 onto HIV-1 gp41 and relevant T20-resistant mechanisms explored by computational study. Current HIV Research. 2012;10:182–194. doi: 10.2174/157016212799937191. [DOI] [PubMed] [Google Scholar]
- 52.Trobridge GD, Wu RA, Beard BC, Chiu SY, Muñoz NM, von Laer D, et al. Protection of stem cell-derived lymphocytes in a primate AIDS gene therapy model after in vivo selection. PLoS ONE. 2009;4:e7693. doi: 10.1371/journal.pone.0007693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zahn RC, Hermann FG, Kim EY, Rett MD, Wolinsky SM, Johnson RP, et al. Efficient entry inhibition of human and nonhuman primate immunodeficiency virus by cell surface-expressed gp41-derived peptides. Gene Ther. 2008;15:1210–1222. doi: 10.1038/gt.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cossarini F, Galli A, Galli L, Bigoloni A, Salpietro S, Vinci C, et al. Immune recovery and T cell subset analysis during effective treatment with maraviroc. J Antimicrob Chemother. 2012 doi: 10.1093/jac/dks216. [Epub ahead of print 2012 Jun 7]. [DOI] [PubMed] [Google Scholar]
- 55.LaBrecque J, Metz M, Lau G, Darkes MC, Wong RS, Bogucki D, et al. HIV-1 entry inhibition by small-molecule CCR5 antagonists: a combined molecular modeling and mutant study using a high-throughput assay. Virology. 2011;413:231–243. doi: 10.1016/j.virol.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 56.Shin N, Solomon K, Zhou N, Wang KH, Garlapati V, Thomas B, et al. Identification and characterization of INCB9471, an allosteric noncompetitive small-molecule antagonist of C-C chemokine receptor 5 with potent inhibitory activity against monocyte migration and HIV-1 infection. J Pharmacol Exp Ther. 2011;338:228–239. doi: 10.1124/jpet.111.179531. [DOI] [PubMed] [Google Scholar]
- 57.Fessel WJ, Anderson B, Follansbee SE, Winters MA, Lewis ST, Weinheimer SP, et al. The efficacy of an anti-CD4 monoclonal antibody for HIV-1 treatment. Antiviral Res. 2011;92:484–487. doi: 10.1016/j.antiviral.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacobson JM, Thompson MA, Lalezari JP, Saag MS, Zingman BS, D'Ambrosio P, et al. Anti-HIV-1 activity of weekly or biweekly treatment with subcutaneous PRO 140, a CCR5 monoclonal antibody. J Infect Dis. 2010;201:1481–1487. doi: 10.1086/652190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duenas-Decamp MJ, O'Connell OJ, Corti D, Zolla-Pazner S, Clapham PR. The W100 pocket on HIV-1 gp120 penetrated by b12 is not a target for other CD4bs monoclonal antibodies. Retrovirology. 2012;9:9. doi: 10.1186/1742-4690-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Da LT, Quan JM, Wu YD. Understanding the binding mode and function of BMS-488043 against HIV-1 viral entry. Proteins. 2011;79:1810–1819. doi: 10.1002/prot.23005. [DOI] [PubMed] [Google Scholar]
- 61.Duong YT, Meadows DC, Srivastava IK, Gervay-Hague J, North TW. Direct inactivation of human immunodeficiency virus type 1 by a novel small-molecule entry inhibitor, DCM205. Antimicrobial Agents & Chemotherapy. 2007;51:1780–1786. doi: 10.1128/AAC.01001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kawai K, Tsuno NH, Kitayama J, Okaji Y, Yazawa K, Asakage M, et al. Epigallocatechin gallate, the main component of tea polyphenol, binds to CD4 and interferes with gp120 binding. Journal of Allergy & Clinical Immunology. 2003;112:951–957. doi: 10.1016/s0091-6749(03)02007-4. [DOI] [PubMed] [Google Scholar]
- 63.Xue J, Gao Y, Hoorelbeke B, Kagiampakis I, Zhao B, Demeler B, et al. The role of individual carbohydrate-binding sites in the function of the potent anti-HIV lectin Griffithsin. Molecular Pharmaceutics. 2012 doi: 10.1021/mp300194b. [Epub ahead of print 2012 Jul 24]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fink RC, Roschek B, Jr, Alberte RS. HIV type-1 entry inhibitors with a new mode of action. Antiviral Chemistry & Chemotherapy. 2009;19:243–255. doi: 10.1177/095632020901900604. [DOI] [PubMed] [Google Scholar]
- 65.Gascoigne NR, Palmer E. Signaling in thymic selection (Review) Curr Opin Immunol. 2011;23:207–212. doi: 10.1016/j.coi.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nepom GT. MHC class II tetramers (Review) J Immunol. 2012;188:2477–2482. doi: 10.4049/jimmunol.1102398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edling AE, Choksi S, Huang Z, Korngold R. An organic CD4 inhibitor reduces the clinical and pathological symptoms of acute experimental allergic encephalomyelitis. J Autoimmun. 2002;18:169–179. doi: 10.1006/jaut.2001.0576. [DOI] [PubMed] [Google Scholar]
- 68.Xiao H, Zhang H, Yu ZY, Zhang L, Yu M, Huang YF, et al. J2 prolongs the corneal allograft survival through inhibition of the CD4+ T cell-mediated response in vivo. Transpl Immunol. 2007;18:130–137. doi: 10.1016/j.trim.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 69.Wilen CB, Wang J, Tilton JC, Miller JC, Kim KA, Rebar EJ, et al. Engineering HIV-resistant human CD4+ T cells with CXCR4-specific zinc-finger nucleases. PLoS Pathogens. 2011;7:e1002020. doi: 10.1371/journal.ppat.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan J, Wang J, Crain K, Fearns C, Kim KA, Hua KL, et al. Zinc-finger nuclease editing of human cxcr4 promotes HIV-1 CD4(+) T cell resistance and enrichment. Mol Ther. 2012;20:849–859. doi: 10.1038/mt.2011.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tachibana K, Hirota S, Iizasa H, Yoshida H, Kawabata K, Kataoka Y, et al. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 1998;393:591–594. doi: 10.1038/31261. [DOI] [PubMed] [Google Scholar]
- 72.Chung SH, Seki K, Choi BI, Kimura KB, Ito A, Fujikado N, et al. CXC chemokine receptor 4 expressed in T cells plays an important role in the development of collagen-induced arthritis. Arthritis Research & Therapy. 2010;12:R188. doi: 10.1186/ar3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laguette N, Benkirane M. How SAMHD1 changes our view of viral restriction (Review) Trends Immunol. 2012;33:26–33. doi: 10.1016/j.it.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan N, Chen ZJ. Intrinsic antiviral immunity (Review) Nat Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 76.Hosseini I, Gabhann FM. Multi-scale modeling of HIV infection in vitro and APOBEC3G-based anti-retroviral therapy. PLoS Computational Biology. 2012;8:e1002371. doi: 10.1371/journal.pcbi.1002371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pham QT, Bouchard A, Grutter MG, Berthoux L. Generation of human TRIM5alpha mutants with high HIV-1 restriction activity. Gene Ther. 2010;17:859–871. doi: 10.1038/gt.2010.40. [DOI] [PubMed] [Google Scholar]
- 78.Martin KL, Johnson M, D'Aquila RT. APOBEC3G complexes decrease human immunodeficiency virus type 1 production. J Virol. 2011;85:9314–9326. doi: 10.1128/JVI.00273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sakuma T, Barry MA, Ikeda Y. Lentiviral vectors: basic to translational (Review) Biochem J. 2012;443:603–618. doi: 10.1042/BJ20120146. [DOI] [PubMed] [Google Scholar]
- 80.Li X, Sodroski J. The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J Virol. 2008;82:11495–11502. doi: 10.1128/JVI.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, Yeager M. Hexagonal assembly of a restricting TRIM5alpha protein. Proc Natl Acad Sci USA. 2011;108:534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, Dekelver RC, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 84.Rosenberg BR, Hamilton CE, Mwangi MM, Dewell S, Papavasiliou FN. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3' UTRs. Nature Structural & Molecular Biology. 2011;18:230–236. doi: 10.1038/nsmb.1975. [Erratum appears in Nat Struct Mol Biol. 2012 Mar;19(3)364] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paz-Yaacov N, Levanon EY, Nevo E, Kinar Y, Harmelin A, Jacob-Hirsch J, et al. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc Natl Acad Sci USA. 2010;107:12174–12179. doi: 10.1073/pnas.1006183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tareen SU, Emerman M. Human Trim5alpha has additional activities that are uncoupled from retroviral capsid recognition. Virology. 2011;409:113–120. doi: 10.1016/j.virol.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anderson J, Akkina R. TRIM5alpharh expression restricts HIV-1 infection in lentiviral vector-transduced CD34+-cell-derived macrophages. Mol Ther. 2005;12:687–696. doi: 10.1016/j.ymthe.2005.07.291. [DOI] [PubMed] [Google Scholar]
- 89.Tyagi M, Bukrinsky M. HIV latency: the major hurdle in HIV eradication. Molecular Medicine. 2012 doi: 10.2119/molmed.2012.00194. [Epub ahead of print 2012 Jun 8] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chun TW, Justement JS, Murray D, Hallahan CW, Maenza J, Collier AC, et al. Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: implications for eradication. AIDS. 2010;24:2803–2808. doi: 10.1097/QAD.0b013e328340a239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luo R, Piovoso MJ, Martinez-Picado J, Zurakowski R. HIV model parameter estimates from interruption trial data including drug efficacy and reservoir dynamics. PLoS ONE. 2012;7:e40198. doi: 10.1371/journal.pone.0040198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gallastegui E, Marshall B, Vidal D, Sanchez-Duffhues G, Collado JA, Alvarez-Fernandez C, et al. Combination of biological screening in a cellular model of viral latency and virtual screening identifies novel compounds that reactivate HIV-1. J Virol. 2012;86:3795–3808. doi: 10.1128/JVI.05972-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xing S, Bullen CK, Shroff NS, Shan L, Yang HC, Manucci JL, et al. Disulfiram reactivates latent HIV-1 in a Bcl-2-transduced primary CD4+ T cell model without inducing global T cell activation. J Virol. 2011;85:6060–6064. doi: 10.1128/JVI.02033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shan L, Deng K, Shroff NS, Durand CM, Rabi SA, Yang HC, et al. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity. 2012;36:491–501. doi: 10.1016/j.immuni.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lafeuillade A. Eliminating the HIV reservoir (Review) Current HIV/AIDS Reports. 2012;9:121–131. doi: 10.1007/s11904-012-0115-y. [DOI] [PubMed] [Google Scholar]
- 97.Forde J, Volpe JM, Ciupe SM. Latently infected cell activation: a way to reduce the size of the HIV reservoir? Bull Math Biol. 2012;74:1651–1672. doi: 10.1007/s11538-012-9729-x. [DOI] [PubMed] [Google Scholar]
- 98.Aubert M, Ryu BY, Banks L, Rawlings DJ, Scharenberg AM, Jerome KR. Successful targeting and disruption of an integrated reporter lentivirus using the engineered homing endonuclease Y2 I-AniI. PLoS ONE [Electronic Resource] 2011;6:e16825. doi: 10.1371/journal.pone.0016825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schiffer JT, Aubert M, Weber N, Mintzer E, Stone D, Jerome KR. Targeted DNA mutagenesis for the cure of chronic viral infections. J Virol. 2012 doi: 10.1128/JVI.00052-12. [Epub ahead of print 2012 Jun 20]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mbonye U, Karn J. Control of HIV latency by epigenetic and non-epigenetic mechanisms. Current HIV Research. 2011;9:554–567. doi: 10.2174/157016211798998736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hu B, Dai B, Wang P. Vaccines delivered by integration-deficient lentiviral vectors targeting dendritic cells induces strong antigen-specific immunity. Vaccine. 2010;28:6675–6683. doi: 10.1016/j.vaccine.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ebina H, Kanemura Y, Suzuki Y, Urata K, Misawa N, Koyanagi Y. Integrase-independent HIV-1 infection is augmented under conditions of DNA damage and produces a viral reservoir. Virology. 2012;427:44–50. doi: 10.1016/j.virol.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 103.Gaur M, Leavitt AD. Mutations in the human immunodeficiency virus type 1 integrase D,D(35)E motif do not eliminate provirus formation. J Virol. 1998;72:4678–4685. doi: 10.1128/jvi.72.6.4678-4685.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Harui A, Suzuki S, Kochanek S, Mitani K. Frequency and stability of chromosomal integration of adenovirus vectors. J Virol. 1999;73:6141–6146. doi: 10.1128/jvi.73.7.6141-6146.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mitani K, Kubo S. Adenovirus as an integrating vector (Review) Current Gene Therapy. 2002;2:135–144. doi: 10.2174/1566523024605591. [DOI] [PubMed] [Google Scholar]
- 106.Dormond E, Kamen AA. Manufacturing of adenovirus vectors: production and purification of helper dependent adenovirus (Review) Methods in Molecular Biology. 2011;737:139–156. doi: 10.1007/978-1-61779-095-9_6. [DOI] [PubMed] [Google Scholar]
- 107.Silva AC, Peixoto C, Lucas T, Kuppers C, Cruz PE, Alves PM, et al. Adenovirus vector production and purification (Review) Current Gene Therapy. 2010;10:437–455. doi: 10.2174/156652310793797694. [DOI] [PubMed] [Google Scholar]
- 108.Duerr A, Huang Y, Buchbinder S, Coombs RW, Sanchez J, Del Rio C, et al. Extended follow-up confirms early vaccine-enhanced risk of HIV acquisition and demonstrates waning effect over time among participants in a randomized trial of recombinant adenovirus HIV vaccine (Step Study) J Infect Dis. 2012;206:258–266. doi: 10.1093/infdis/jis342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Qureshi H, Ma ZM, Huang Y, Hodge G, Thomas MA, DiPasquale J, et al. Low-dose penile SIVmac251 exposure of rhesus macaques infected with adenovirus type 5 (Ad5) and then immunized with a replication-defective Ad5-based SIV gag/pol/nef vaccine recapitulates the results of the phase IIb step trial of a similar HIV-1 vaccine. J Virol. 2012;86:2239–2250. doi: 10.1128/JVI.06175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hawkins CJ. TRAIL and malignant glioma (Review) Vitamins & Hormones. 2004;67:427–452. doi: 10.1016/S0083-6729(04)67022-1. [DOI] [PubMed] [Google Scholar]
- 111.Schuhmann NK, Pozzoli O, Sallach J, Huber A, Avitabile D, Perabo L, et al. Gene transfer into human cord blood-derived CD34(+) cells by adeno-associated viral vectors. Exp Hematol. 2010;38:707–717. doi: 10.1016/j.exphem.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 112.Srivastava A. Gene delivery to human and murine primitive hematopoietic stem and progenitor cells by AAV2 vectors. Methods in Molecular Biology. 2004;246:245–254. doi: 10.1385/1-59259-650-9:245. [DOI] [PubMed] [Google Scholar]
- 113.Veldwijk MR, Sellner L, Stiefelhagen M, Kleinschmidt JA, Laufs S, Topaly J, et al. Pseudotyped recombinant adeno-associated viral vectors mediate efficient gene transfer into primary human CD34(+) peripheral blood progenitor cells. Cytotherapy. 2010;12:107–112. doi: 10.3109/14653240903348293. [DOI] [PubMed] [Google Scholar]
- 114.Handel EM, Gellhaus K, Khan K, Bednarski C, Cornu TI, Muller-Lerch F, et al. Versatile and efficient genome editing in human cells by combining zinc-finger nucleases with adeno-associated viral vectors. Hum Gene Ther. 2012;23:321–329. doi: 10.1089/hum.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li H, Lasaro MO, Jia B, Lin SW, Haut LH, High KA, et al. Capsid-specific T-cell responses to natural infections with adeno-associated viruses in humans differ from those of nonhuman primates. Mol Ther. 2011;19:2021–2030. doi: 10.1038/mt.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhou J, Li H, Zhang J, Piotr S, Rossi J. Development of cell-type specific anti-HIV gp120 aptamers for siRNA delivery. Journal of Visualized Experiments. 2011 doi: 10.3791/2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhou J, Neff CP, Liu X, Zhang J, Li H, Smith DD, et al. Systemic administration of combinatorial dsiRNAs via nanoparticles efficiently suppresses HIV-1 infection in humanized mice. Mol Ther. 2011;19:2228–2238. doi: 10.1038/mt.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects (Review) Chemistry & Biology. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Barber GN. Cytoplasmic DNA innate immune pathways (Review) Immunol Rev. 2011;243:99–108. doi: 10.1111/j.1600-065X.2011.01051.x. [DOI] [PubMed] [Google Scholar]
- 120.Kariko K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]