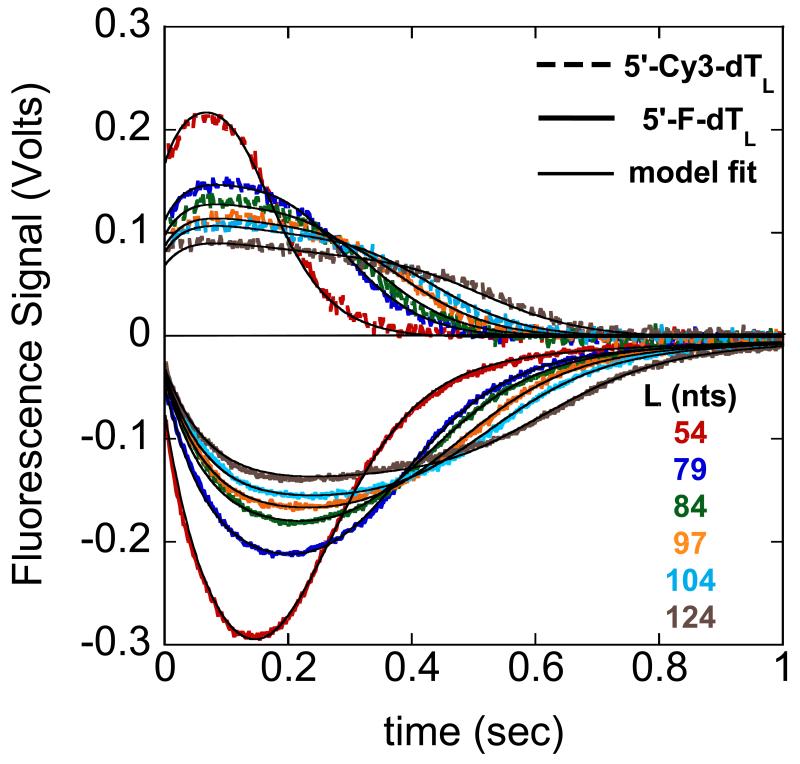
Figure 3.2.
UvrD translocation along single strand DNA labeled at the 5′-end with either fluorescein (F) or Cy3. UvrD is pre-incubated with excess DNA then rapidly mixed in the stopped-flow with ATP, MgCl2, and heparin to initiate translocation (final conditions: 10mM Tris-HCl, pH 8.3, 20mM NaCl, 20%(v/v) glycerol, 25nM UvrD, 50nM DNA, 0.5mM ATP, 2mM MgCl2, 4mg/ml heparin at 25°C). Heparin serves as a protein trap for UvrD, preventing UvrD from rebinding to the DNA, reinitiating translocation. The resulting fluorescence time courses for fluorescein (solid lines) and Cy3 (dashed lines) labeled single strand DNA of differing lengths is shown. The single strand DNA is composed of deoxythymidylates to avoid formation of secondary structures that may have an effect on translocation. The black curves are a global fit to both data sets using a n-step sequential model (27) yielding a value of m*kt = (193 ± 1) nucleotides/second.