Figure 3.
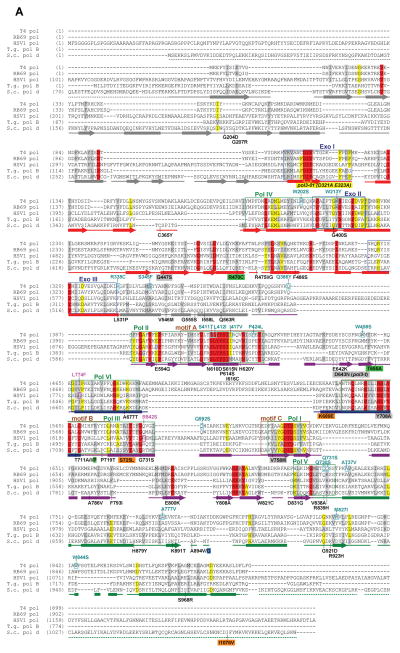
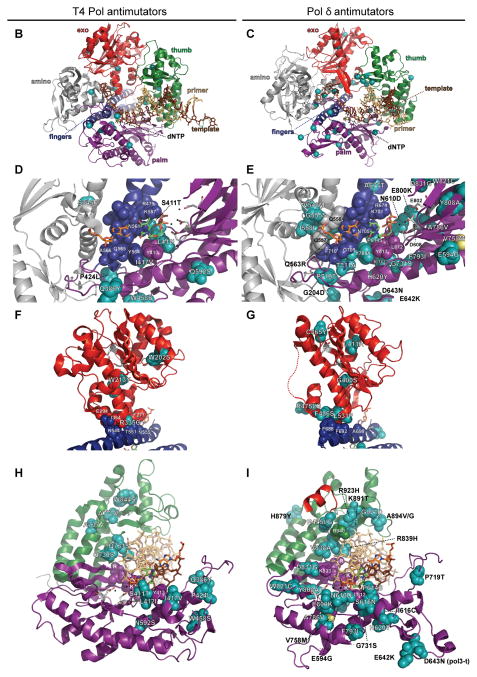
B-family polymerase antimutators. (A) Aligned sequences of five B-family DNA polymerases are shown: bacteriophage T4 (T4 Pol), bacteriophage RB69 (RB69 pol), Herpes Simplex Virus 1 (HSV1 pol), Thermococcus gorgonarius (T.g. pol B), and Saccharomyces cerevisiae (S.c. pol d). Secondary structural elements of yeast Pol δ (Swan et al., 2009) are indicated below the alignment and color coded to depict their locations in the amino (gray), exo (red), palm (purple), fingers (blue) and thumb (green) domains of the protein: rectangles, α-helices; arrows, β-strands; solid lines, loops; dashed lines, structure not solved. Highlighted amino-acid residues in the alignment correspond to the following: red, absolutely conserved; yellow, identical in majority of sequences; gray, similar in majority of sequences. Conserved polymerase and exonuclease motifs are indicated by colored frames: blue, Exo motifs; green, Pol motifs (Wang et al., 1989); brown, motifs A, B, C (Delarue et al., 1990). Amino-acid substitutions of interest are placed above or below the alignment at the relevant position, highlighted according to the following scheme: aqua, T4 Pol antimutators (Reha-Krantz, 1995a; Reha-Krantz and Wong, 1996; Reha-Krantz, 2010); magenta, HSV1 Pol antimutators (Hall et al., 1984; Gibbs et al., 1988; Hwang et al., 2004; Tian et al., 2009); yellow, yeast pol3-01; no highlight, antimutators isolated as suppressors of the pol3-01 mutator phenotype; green, antimutators isolated as suppressors of the pol3-D407A mutator phenotype; orange, antimutators isolated as suppressors of the pol3-F406A mutator phenotype (three substitutions in the same mutant); blue, antimutator isolated as a suppressor of the pol3-D463A mutator phenotype; gray, previously reported antimutators in Pol δ (pol3-t (D643N), G447S, Y708A, and V758M) (Tran et al., 1999a; Hadjimarcou et al., 2001; Pavlov et al., 2004; Li et al., 2005). The antimutators affecting pol3-01, pol3-D407A, pol3-F406A, and pol3-D463A are from Herr et al. (Herr et al., 2011). (B-I) Locations of antimutator amino-acid substitutions in T4 Pol [B, D, F, and H; mapped onto RB69Pol (Franklin et al., 2001), Protein Data Bank accession code 1IG9] and Pol δ [C, E, G, and I; mapped onto Pol δ (Swan et al., 2009), PDB accession code 3IAY]. The proteins are shown as schematic diagrams of the α-carbon backbone with α-carbons or amino acids of interest depicted as space-filling spheres. Wild type amino-acid residues corresponding to antimutator mutations are shown as teal spheres and labeled to indicate the antimutator substitutions. Structural domains are color coded as in the alignment in panel A. The incoming dNTPs are denoted by green CPK sticks and the template nucleotides by orange CPK sticks. Active-site carboxylate side chains are gray CPK sticks coming out of the purple (palm) or red (exo) ribbons. Metal ions are small black spheres. Important non-mutated residues proximal to the antimutator substitutions are also shown as space-filling spheres and color coded according to domain. (B and C) Distribution of antimutator substitutions. The α-carbons of wild type residues corresponding to antimutators are shown as teal spheres. (D and E) Antimutator substitutions near the polymerase active site. The fingers and thumb domains have been removed for clarity. T4 Pol S345 and Pol δ residues V546, G555, I558, and Q563 from the amino domain are in three closely associated α-helices that bind the template and buttress the fingers domain. The antimutator substitution (V758M) that conferred PAA-resistance to a Pol δ-L612M strain is shown in yellow. (F and G) Antimutator substitutions in the exo domain. All T4 Pol antimutator variants occurred in the context of a functional exonuclease, unlike Pol δ antimutator polymerases, which carried D321A and D323A substitutions that inactivate the exonuclease. The dotted red line in (G) corresponds to a missing loop in the Pol δ structure (amino acids 491–496). (H and I) Antimutator substitutions in the DNA binding track. The primer (gold) and template (brown) are held by a series of interactions along the DNA minor groove. Minor-groove ‘sensing’ residues in the KKRYA motif of the palm domain (T4 Pol K702, K703, R704; Pol δ K813, K814 and R815) are shown as space-filling spheres labeled K and R. Unpaired 5′ nucleotides of the template along with the amino, exo, and fingers domains have been removed for clarity. A color version of this figure is available online.