Figure 3. Structural Elements of IES Regions and Transposon-Derived Genes or Systems.
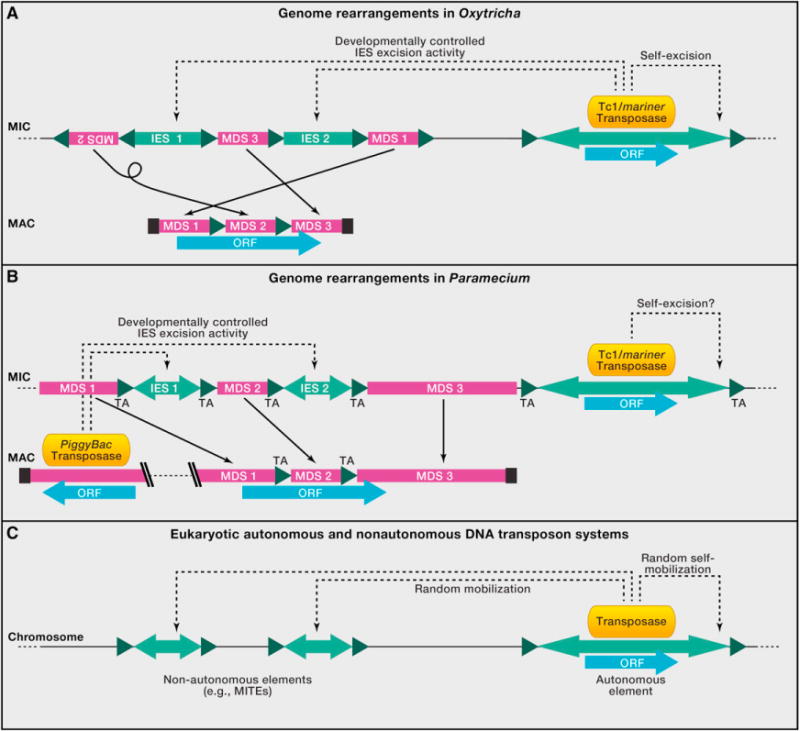
Magenta boxes, macronuclear-destined sequence (MDS); light green, internal eliminated sequences (IES) or transposable elements. Inverted light green arrowheads indicate inverted repeats at ends of deleted sequences. Dark green triangles indicate short direct repeats (pointer sequences). Blue bars indicate open reading frames (ORFs). Solid thin arrows show correspondence between DNA sequences in the MIC and MAC, and black boxes indicate telomeres on MAC chromosomes.
(A) Shown on the left is a schematic scrambled gene in Oxytricha or Stylonychia. In Oxytricha, thousands of MIC-encoded Tc1/mariner transposases (orange) likely participate in removal of IESs (indicated by dotted arrows) as well as themselves (Nowacki et al., 2009).
(B) In oligohymenophorean ciliates, represented by Paramecium, IES excision requires a domesticated PiggyBac transposase encoded in the MAC. Paramecium has only TA dinucleotides as pointers and also flanking Tc1/mariner transposons in the MIC (Arnaiz et al., 2012). Euplotes crassus, a spirotrich, similarly has nonscrambled IESs with TA dinucleotides flanking both IESs and Tec transposons (Jacobs and Klobutcher, 1996). Tetrahymena differs from Paramecium in having mostly imprecise excision of longer intergenic IESs, with some precisely excised IESs flanked by TTAA repeats.
(C) Schematic illustration of eukaryotic nonautonomous and autonomous DNA transposons. The autonomous elements (right) encode transposases that can mobilize truncated and simplified nonautonomous elements (left) throughout the genome. The structure of these elements mirrors the structure of ciliate IESs (nonautonomous elements) and their controlling transposons (autonomous elements), most likely reflecting their evolutionary origins and development (see Klobutcher and Herrick, 1997).
