Figure 4. Two Models of RNA-Mediated Genome Rearrangements in Ciliates.
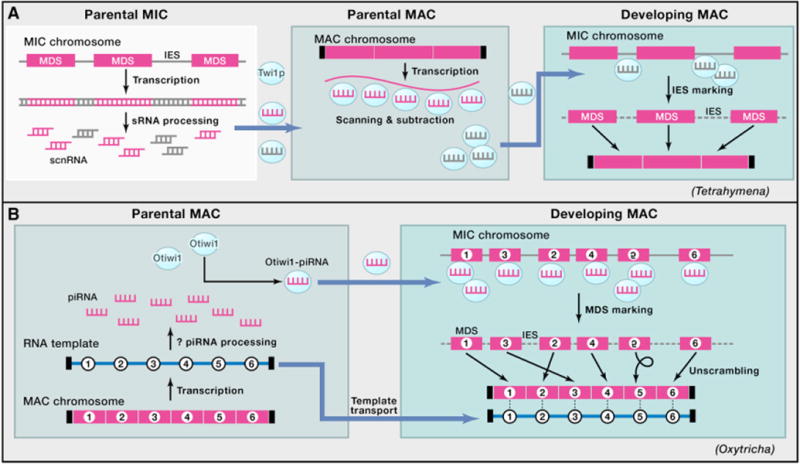
(A) In Tetrahymena, bidirectional transcription of the MIC genome produces double-stranded RNA, which is processed into scnRNA duplexes. The Piwi protein Twi1p loads scnRNAs in the cytoplasm and transports them into the parental MAC to scan the somatic transcriptome. This process enriches for scnRNAs that do not pair with homologous sequences from the parental MAC. The MIC-limited scnRNAs are then transported into the developing MAC, where they recognize and mark IES regionsonMIC chromosomes. IES excision and telomere addition produce mature MAC chromosomes. In both panels, magenta rectangles denote MDSs, and combs indicate sRNA.
(B) In Oxytricha, transcription of either strand of gene-sized chromosomes in the parental MAC produces telomere-containing template RNAs (blue numbered line) during conjugation. Either these template RNAs or other long noncoding RNAs are processed into 27 nt piRNAs. These form a complex with the Piwi protein Otiwi1 that transports them into the newly developing MAC, where the piRNAs recognize and mark the MDS portions of the MIC chromosome that are retained during genome rearrangement. The maternal template RNAs are also transported to the developing MAC, where they guide correct MDS ordering of numbered segments 1–6 (including inversion of segment 5) and DNA repair at recombination junctions to produce mature somatic chromosomes that are capped with short telomeres (small vertical black bars).
