Figure 4. Phylogenetic analysis of TFB protein families in selected eukaryotic species.
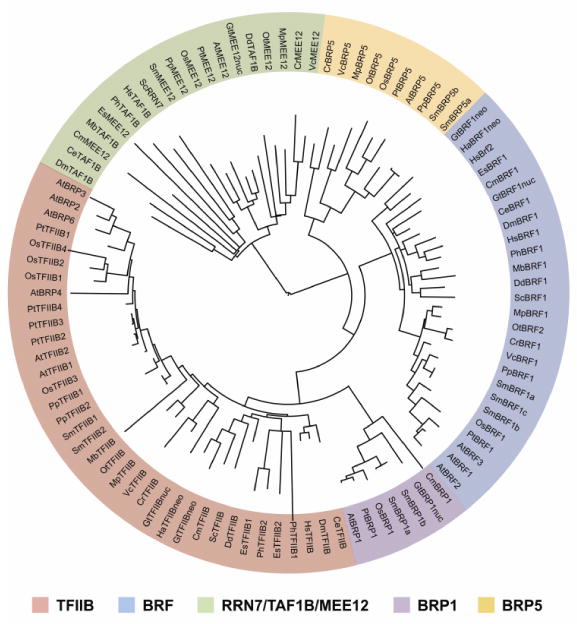
The phylogenetic tree was created by comparing TFIIB homology domains that consist of the zinc ribbon, linker, and cyclin fold domains in each of the selected eukaryotic organisms. Saccharomyces cerevisiae (Sc), Monosiga brevicollis (Mb), Dictyostelium discoideum (Dd), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Homo sapiens (Hs), Phaeodactylum tricornutum (Ph), Ectocarpus siliculosus (Es), Hemiselmis andersenii (Ha), Guillardia theta (Gt), Cyanidioschyzon merolae (Cm), Ostreococcus tauri (Ot), Volvox carteri (Vc), Chlamydomonas reinhardtii (Cr), Physcomitrella patens (Pp), Selaginella moellendorffii (Sm), Oryza sativa (Os), Populus trichocarpa (Pt), Arabidopsis thaliana (At). Each major TFB protein subfamily is shaded by a unique color. The TFB subfamilies include TFIIB, Brf, Rrn7/TAF1B/MEE12, Brp1, and Brp5. Nuclear genome, nuc; Nucleomorph genome, neo. The tree was visualized with iTOL80,81.
