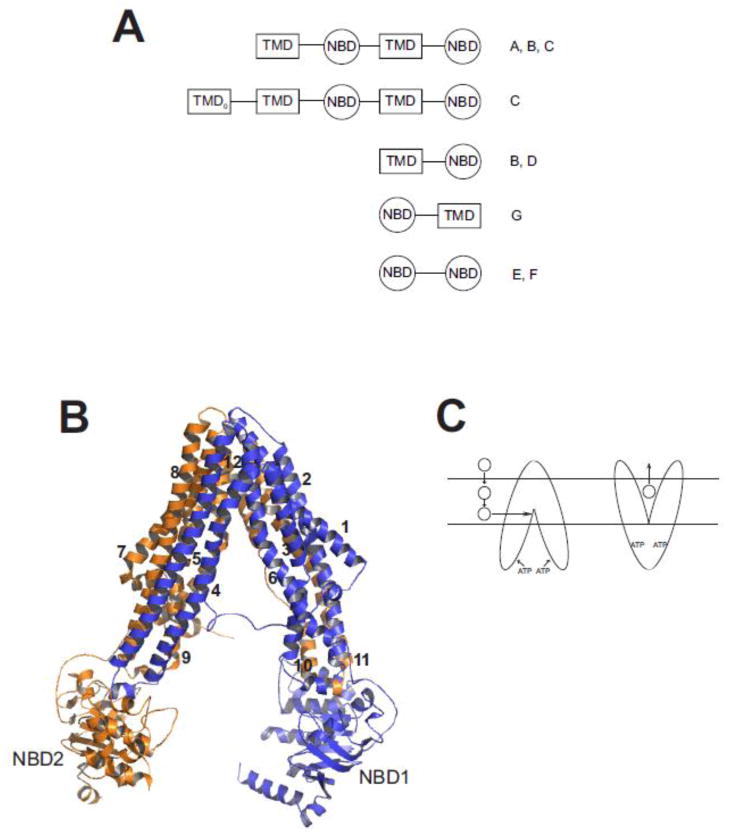
Fig. 1. Structure of ABC multidrug transporters.
A. Predicted domain arrangement of ABC transporters found in the human genome. Shown are the arrangement of transmembrane domains (TMD) and nucleotide binding domains (NBD) found in human ABC transporters. The TMD0 domain is found in some members of the ABCC sub-family. Letters on to the right of the cartoon designate ABC sub-families in which that predicted domain topology is found. Figure adapted from [41] and [35]. B. Homology model of SMDR2. SMDR2 was modeled against C. elegans Pgp (pdb 4F4C; [55]) using ESyPred3D (http://www.fundp.ac.be/sciences/biologie/urbm/bioinfo/esypred/) [113]. Residues at the N-terminus (1–4), linker (609–656), and C-terminus (1243–1255) were not included in the final model. The figure was generated using the program PyMOL (http://www.pymol.org/). NBD1 and NBD2 designate the nucleotide binding domains, and the numbers 1–12 indicate the helical transmembrane-spanning segments. Domain 1 is in blue; domain 2 is in orange. C. Model for binding and transport of substrate by Pgp. Substrate (circle) partitions from outside the cell into the inner leaflet of the lipid bilayer, where, according to the hydrophobic “vacuum cleaner” model, it enters the drug-binding pocket of Pgp. ATP binding to the NBDs produces a conformational change that results in presentation of the substrate to the outer leaflet or extracellular space.