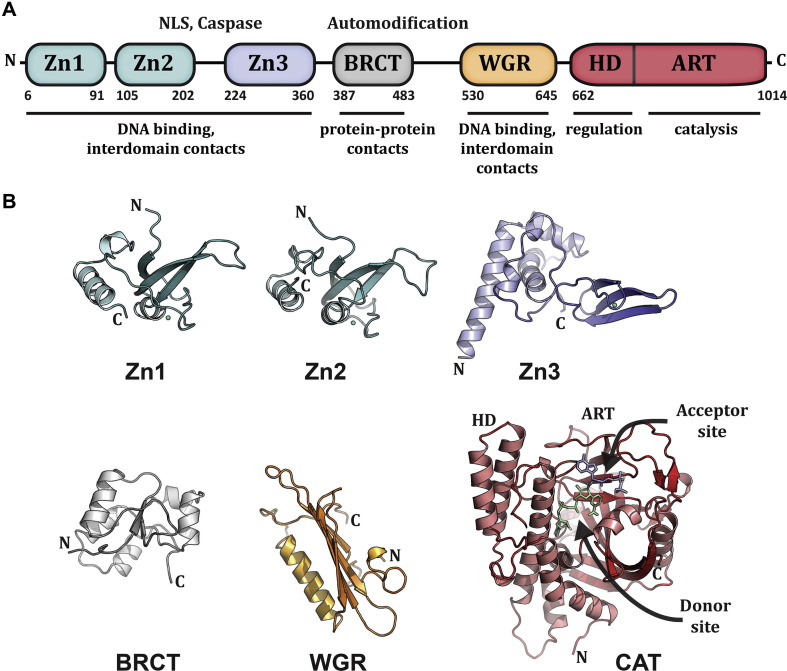
Fig. 4.
PARP1 modular domain structure. (A). Schematic representation of human PARP1 domains. The amino acid numbering for human domain boundaries are noted. Homologous DNA-binding zinc finger domains, Zn1 and Zn2, are located at the N-terminus of PARP1. These domains are followed by a linker region containing a bipartite nuclear localization signal (NLS) and a caspase three cleavage site (Caspase). This regulatory region is followed by a third zinc finger domain (Zn3), which has a distinct structure and function from that of Zn1 and Zn2. A BRCA C-terminus (BRCT) fold is located within the region of PARP1 that is primarily targeted for automodification. The C-terminal end of PARP1 contains a WGR domain, named after a conserved Trp-Gly-Arg sequence, and the catalytic domain, which is composed of an α-helical subdomain (HD) and an ADP-ribosyl transferase subdomain (ART). The known functions for the individual domains are noted beneath the schematic. (B). Crystal and/or NMR structures have been determined for each of the PARP1 domains in the absence of DNA: the NMR structures for the homologous Zn1 and Zn2 domains [PDB: 2DMJ and 2CS2; see also reference (Eustermann et al., 2011)], the NMR structure of the Zn3 domain is shown [PDB: 2JVN; see also (Langelier et al., 2008) for crystal structure determination], the NMR structure of the BRCT fold [PDB: 2COK; see also (Loeffler et al., 2011), the NMR structure of the WGR domain (PDB: 2CR9), and the crystal structure of the catalytic domain (PDB: 1A26) (Ruf et al., 1996)]. Catalytic subdomains are labeled (HD and ART). The crystal structure was determined in the presence of an NAD+ analog that has defined the proposed “acceptor site” for poly(ADP-ribose) formation. The NAD+ binding site is modeled in this figure based on the structure of an ADP-ribosylating toxin structure (Bell and Eisenberg 1996). Reprinted from (Langelier and Pascal 2013) with permission from Elsevier.