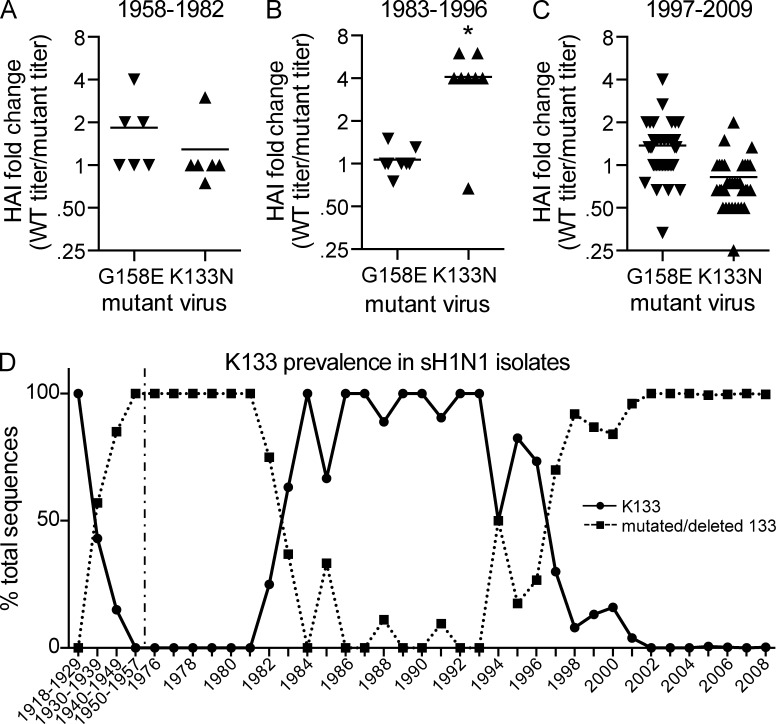
Figure 3.
pH1N1 antibody responses in humans born between 1983–1996 are dominated against region of HA involving K133. (A–C) HAI assays were completed using viruses possessing either wild-type A/California/07/2009 HA or A/California/07/2009 HA with G158E or K133N mutations. HAI assays were performed with sera isolated from humans 9–31 d after onset of pH1N1 symptoms. Humans were naturally infected with pH1N1 (PCR-verified). HAI titers are shown in Table S3. Fold change for each mutant virus (WT HAI titer/mutant HAI titer) is shown here. Each triangle represents an individual sera sample, and the mean is indicated with a line. Individuals are separated based on year of birth. 7 of 8 sera samples from individuals born between 1983 and 1996 had reduced titers to the K133N mutant, whereas only 2 sera samples from the other time periods (n = 46) had reduced titers to the K133N mutant (K133-specificity of sera from 1983–1996 is significantly different compared with sera from other time periods; Fisher’s exact test; *, P < 0.001). HAI data are representative of three independent experiments. (D) Timeline depicting the percentage of sH1N1 isolates possessing Lys (solid line) or a mutation/deletion (dotted line) at position 133 of HA based on sequences contained in the NCBI database. In total, 7,045 sequences resulting from unique isolates were aligned using the program MUSCLE to yield K133 prevalence on a yearly basis. Isolates from 1918–1957 were grouped due to low number of sequences from these years. sH1N1 viruses did not circulate between 1958 and 1976, and this is indicated by a dashed vertical line.