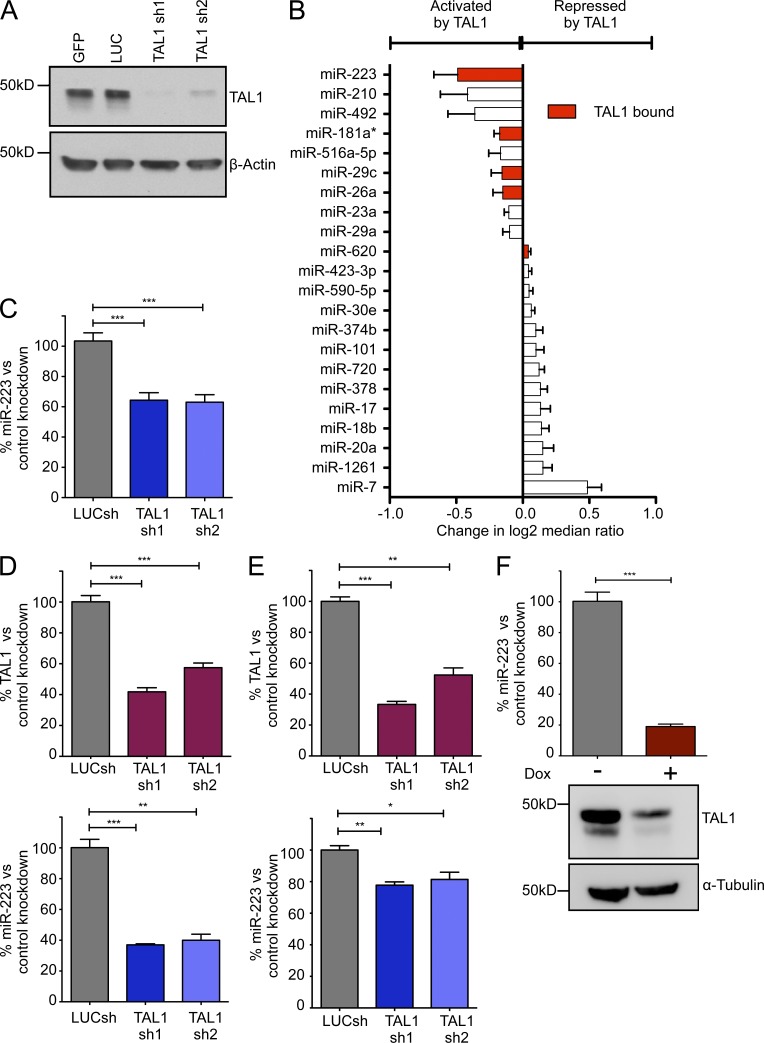
Figure 1.
TAL1 regulates a select number of miRNAs in T-ALL cells. (A) TAL1 was silenced by lentiviral knockdown in Jurkat cells and protein expression was assessed by Western blot. Two independent shRNAs were compared with control shRNAs targeting GFP or luciferase (LUC). (B) Global miRNA expression profiling with the Exiqon miRCURY LNA platform after shRNA knockdown of TAL1 in Jurkat cells. Horizontal bars show the mean change in log2 median ratios (Hy3/Hy5) ± SD after TAL1 knockdown in duplicate experiments with shRNAs 1 and 2 compared with control shRNAs targeting GFP or luciferase. Only miRNAs with a P value ≤ 0.05 by two-tailed Student’s t test are shown. Red bars represent miRNAs bound by TAL1 in Jurkat cells as determined by ChIP-seq analysis. (C) Quantitative RT-PCR analysis of miR-223 expression normalized to RNU5A in Jurkat cells after TAL1 knockdown with shRNAs 1 and 2 compared with a LUC control shRNA. Bars represent mean ± SEM from three independent experiments performed in triplicate. TAL1 was silenced in RPMI-8402 (D) and CCRF-CEM (E) cells and miR-223 expression was determined by qRT-PCR. (top) TAL1 expression normalized to GAPDH and bottom panel shows miR-223 expression normalized to RNU5A. Bars represent the mean ± SEM of two experiments performed in triplicate. (F) miR-223 expression was assessed by qRT-PCR in a doxycycline-inducible TAL1 knockdown system developed in Jurkat cells (Jurkat S1C1) for 48 h with (+) or without (−) the addition of doxycycline (Dox). Bars represent the mean ± SEM of two experiments performed in triplicate. Western blot (bottom) shows TAL1 protein expression after the addition of doxycycline. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 by two-tailed Student’s t test.