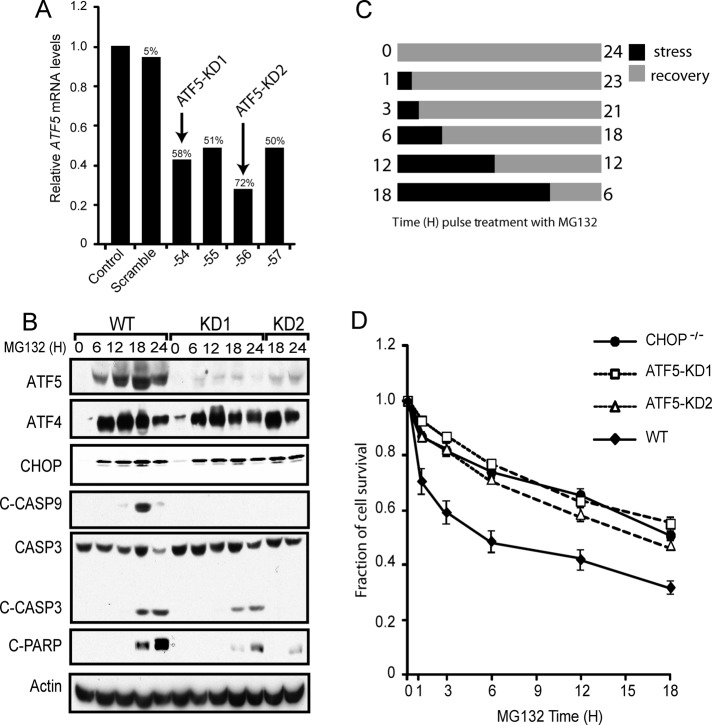
FIGURE 5:
ATF5 facilitates apoptosis in response to proteasome inhibition. (A) RNA was isolated from MEF cells stably expressing a scrambled shRNA or an ATF5-targeted shRNA, and ATF5 mRNA levels were determined by qPCR relative to non-shRNA–expressing control. The ID number indicates the last two digits for the shRNA. The percentages of ATF5 knockdown were determined relative to a non-shRNA–expressing control. (B) WT, ATF5-KD1, and ATF5-KD2 MEF cells were treated with MG132 for the indicated time points. Levels of ATF4, CHOP, ATF5, cleaved caspase-9, cleaved caspase-3, cleaved PARP, and actin proteins were measured by immunoblot analysis using specific antibodies. (C) Cell treatment regimen for the data shown in D. Cells were cultured with 1 μM MG132 for 0, 1, 3, 6, 12, or 18 h as indicated by the black bars, and after the stress treatment, cells were cultured in the absence of MG132, as indicated by the gray bars, for a total of 24 h. (D) Measurements of survival of WT, CHOP−/−, ATF5-KD1, and ATF5-KD2 MEF cells were determined by the MTT assay, and the percentage of cell survival is normalized to the untreated controls for each cell type.